Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers and the second leading cause of cancer death worldwide [1]. Despite the therapeutic advances in HCC in the past few decades, the mortality rate of HCC is still high [2]. Chronic hepatitis B (HBV) and hepatitis C (HCV) infection were the major etiological risk factors of HCCs [3]. HCV is a sense-strand RNA virus and is unable to integrate into the host genome [4] and HCV infection represents the main risk factor of primary HCC [5]. Although a previous study showed that several proteins (such as COX-2) were associated with decreased overall and disease-free survival [6], and HCV encoded proteins (such as E1, E2, P7, NS2 and NS3) could also promote HCC formation by activating oncogenic molecular pathways in vivo [7–9], the underlying mechanisms of HCV-induced hepatocarcinogenesis remain largely unclear.
LncRNAs are a class of non-coding RNAs longer than 200 nucleotides with little or no protein-coding potential [10]. Previous studies had reported that lncRNA expression was frequently dysregulated and could contribute to the progress of various human cancer types, including prostate [11], breast [12], colon [13] and liver cancers [14]. For example, Fan et al. revealed that more than 1000 lncRNA transcripts were significantly differentially expressed in HBV-related HCC patients [15]. Mechanically, lncRNAs could regulate protein-coding genes’ expression at epigenetic, transcriptional, and post-translational levels [16]. One of the most well-known mechanisms was acting as miRNAs sponges by interacting with miRNAs [17]. However, the potential roles of an enormous number of lncRNAs in HCC, especially in HCV-positive HCC, remains to be further elucidated.
In this study, we analyzed the public dataset GSE17856 to identify differentially expressed lncRNAs (DElncs) and mRNAs in HCV-related HCC patients. Next, a series of bioinformatics analyses (including GO, KEGG and co-expression analysis) were performed to explore potential roles of DElncs. The present study aimed to provide useful information to identify novel lncRNAs as biomarkers for HCV-related HCC.
Material and methods
Microarray data
Microarray data were obtained from the study by Tsuchiya et al. [18], which is referenced in the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) under accession number GSE17856. The platform is GPL6480 Agilent-014850 Whole Human Genome Microarray 4x44K G4112F Array. A total of 44 non-tumoral liver and 43 tumors sample were obtained from 47 subjects with HCV-associated HCC. LncRNAs having fold changes ≥ 2 and p-values < 0.05 were selected as of significantly differential expression.
LncRNA classification pipeline
To evaluate the lncRNA expression pattern in microarray data, we applied a pipeline described by Zhang et al. [19]. Briefly, the probe set ID was mapped to the NetAffx Annotation Files (HG-U133 Plus 2.0, Annotations, CSV format, release 31, 08/23/10). The annotations included the probe set ID, gene symbol, gene title, ensemble gene ID, Refseq transcript ID and other informative items for the specific probe sets. The probe ID-centric gene expression data were joined with the annotation files on the probe set ID. Secondly, the probe sets which were assigned with a Refseq transcript ID and/or Ensemble gene ID in the, NetAffx annotations were extracted. For the probe sets with Refseq IDs, we only retained those labeled as “NR_” (NR indicates non-coding, RNA in the Refseq database). For the probe sets with Ensemble gene IDs, we only retained those annotated with “lncRNA”, “processed, transcripts”, “non-coding” or “misc_RNA” in Ensemble annotations (accessible at the UCSC genome browser: http://www.genome., ucsc.edu/). Then, we filtered the probe sets obtained in step 2 by filtering out rRNAs, microRNAs and other short RNAs, including tRNAs, snRNAs and snoRNAs. Finally, 2448 annotated lncRNA transcripts with corresponding Affymetrix probe IDs were generated.
Identification of DElncs
A t-test [20] in the limma [21] package, in R [22] was used to identify genes which were differentially expressed between normal and HCV-positive HCC samples. The threshold for the DEGs was set as corrected pvalue < 0.05 and | log2 fold change, (FC) | ≥ 1.
GO and KEGG pathway analysis
To identify functions of DEGs in HCC, we performed GO function enrichment analysis in three functional ontologies: biological process (BP), cellular component (CC) and molecular function (MF). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was also performed to identify pathways enriched in HCC using the DAVID system (https://david.ncifcrf.gov/). The p-value was calculated by hypergeometric distribution and a pathway with p < 0.05 was considered as significant.
Co-expression network construction and analysis
In this study, the Pearson correlation coefficient of DEG-lncRNA pairs was calculated according to the expression value of them. The co-expressed DEG-lncRNA pairs with the absolute value of Pearson correlation coefficient ≥ 0.5 were selected and the co-expression network was established using Cytoscape software.
Statistical analysis
The numerical data were presented as mean ± standard deviation (SD) of at least three determinations. Statistical comparisons between groups of normalized data were performed using the t-test or Mann-Whitney U-test according to the test condition. A value of p < 0.05 was considered to indicate statistical significance with a 95% confidence level.
Results
Screening the DElncs in the HCV-positive hepatocellular carcinoma
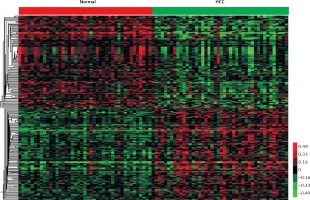
In the present study, the public dataset GSE17856 was analyzed to identify differentially expressed lncRNAs. A total of 44 non-tumoral liver and 43 tumor samples were obtained from 47 subjects with HCV-associated HCC. LncRNAs having fold changes ≥ 2 and p-values < 0.05 were selected as of significantly differential expression. In total, 454 transcripts were observed to be expressed differentially compared to the normal liver tissues, including 256 upregulated transcripts and 198 downregulated transcripts. Hierarchical clustering showed systematic variations in the expression of lncRNAs in the HCV-positive HCC samples (Figure 1).
Figure 1
Hierarchical clustering of systematic variations in the expression of lncRNAs in the HCV-positive HCC samples. We identified 256 upregulated lncRNAs and 198 downregulated lncRNAs in HCV-positive HCC compared to the normal liver tissues. Red indicates high relative expression and green indicates low relative expression. Normal represents nontumoral liver and HCC represents tumor samples which were obtained from subjects with HCV-associated HCC
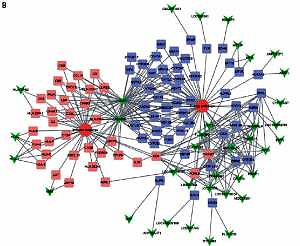
Co-expression network analysis
Here, we constructed up- and down-regulated lncRNA-mRNA co-expressed networks to reveal the potential roles of lncRNAs in HCV-positive HCC. The co-expressed lncRNA-mRNA pairs with the absolute value of the Pearson correlation coefficient ≥ 0.5 were selected for network construction. As shown in Figure 2, a total of 125 lncRNAs and 1469 DEGs were included in the up-regulated lncRNA mediated co-expression network. In this network, we found that LOC341056, CCT6P1, PTTG3P, LOC643387, CXADRP2, LOC100128098, LOC100133920 played key roles and co-expressed with more than 100 DEGs. Meanwhile, we observed that the up-regulated lncRNA mediated co-expression network contained 67 lncRNAs and 558 DEGs (Figure 2 A). Twelve lncRNAs (FLJ35390, MGC45922, LOC92249, MTMR9LP, LOC440461, LOC92659, LOC100130557, LINC00304, RRP7B, FAM201A, PYY2, PIN1P1) co-expressed with more than 100 DEGs in the down-regulated lncRNA mediated network (Figure 2 B).
Functional annotation of differentially expressed lncRNAs
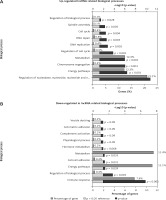
Based on co-expression networks, we performed GO and KEGG analysis for differentially expressed lncRNAs using the set of co-expressed mRNAs. According to the GO analysis, we found that up-regulated lncRNAs were mainly involved in regulating metabolism, energy pathways, regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism, regulation of the cell cycle, amino acid and derivative metabolism, cell cycle, DNA repair, and DNA replication (Figure 3 A). Meanwhile, we observed that the downregulated lncRNAs were enriched in regulating metabolism, energy pathways, immune response, regulation of biological process, and cell-cell adhesion (Figure 3 B).
Identification of key lncRNAs in regulating HCV-positive HCC development
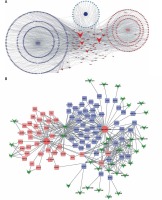
For further explore the key lncRNAs in regulating key biological processes in HCV-positive HCC development, we constructed lncRNA-mRNA-biological processes. We observed that LOC341056, CCT6P1, PTTG3P, LOC643387, LOC100133920 were mainly involved in regulating metabolism and regulation of nucleobase and cell proliferation processes were most significant in the upregulated network (Figure 4 A). In the down-regulated network, C3P1 and C22orf45 played crucial roles in regulating immune response and energy pathways (Figure 4 B).
Figure 4
Identification of key lncRNAs in regulating HCV-positive HCC development. We constructed upregulated lncRNA-mRNA-biological processes. LOC341056, CCT6P1, PTTG3P, LOC643387, and LOC100133920 were involved in regulating metabolism, regulation of nucleobase and cell proliferation processes (A). Down-regulated lncRNAs (C3P1 and C22orf45) were involved in regulating immune response and energy pathways (B). Triangle nodes, lncRNAs; square nodes, lncRNAs co-expressed genes; circle, biological processes
Key lncRNAs were associated with hepatocellular carcinoma prognosis
Furthermore, we evaluated the possible prognostic value of LOC341056, CCT6P1, PTTG3P, LOC643387, LOC100133920, C3P1 and C22orf45 in HCC using TCGA RNA-seq data. As shown in Figures 5 A–D, according to Kaplan-Meier analysis, we found that higher expression levels of LOC643387, PTTG3P, LOC341056, and CCT6P1 were associated with shorter survival time in the TCGA dataset. However, we found that C3P1-high (Figure 5 F) and C22orf45-high (Figure 5 E) patients showed higher survival rates compared to C3P1-and C22orf45-low patients.
Figure 5
Key lncRNAs were associated with hepatocellular carcinoma prognosis. According to Kaplan-Meier analysis, we found that higher expression levels of LOC643387 (A), PTTG3P (B), LOC341056 (C), and CCT6P1 (D) were associated with shorter survival time in TCGA dataset. We found that C22orf45- (E) and C3P1- (F) high patients showed higher survival rates compared to C3P1- and C22orf45-low patients
Significance was defined as p < 0.05 (*p < 0.05; **p < 0.01; ***p < 0.001).

Discussion
Hepatocellular carcinoma is one of the most common cancers worldwide [1]. Hepatitis C infection was one of the major etiological risk factors of HCCs [8]. However, the underlying mechanisms of HCV-induced hepatocarcinogenesis remain largely unclear. LncRNAs were a class of non-coding RNAs of more than 200 nucleotides in length [10]. A previous study demonstrated the key roles of lncRNAs in various human cancer types, including liver cancers [14]. In this study, we analyzed the public dataset GSE17856 and found that 454 lncRNAs (including 256 upregulated lncRNAs and 198 downregulated lncRNAs) were differentially expressed compared to the normal liver tissues.
Recently, emerging studies have shown that lncRNAs were also associated with the progression of HCC. For example, authors [23–25] identified differently expressed lncRNAs in HCV-related HCC using RNA sequencing and expression profile analysis. Zhu et al. reported that LINC00052 upregulated EPB41L3 to inhibit migration and invasion of hepatocellular carcinoma by binding miR-452-5p [26]. LncRNA TSLNC8 was reported as a tumor suppressor by inactivating the IL-6/STAT3 signaling pathway [27]. However, very few reports have focused on the molecular function of lncRNAs in HCV-positive HCC. In this study, we performed bioinformatics analysis to explore the potential roles of lncRNAs in regulating HCV-positive HCC development. We first constructed dysregulated lncRNA-mRNA co-expressed networks. We found 125 up-regulated lncRNAs co-expressed with 1469 DEGs and 67 down-regulated lncRNAs co-expressed with 558 DEGs. Bioinformatics analysis showed that dysregulated lncRNAs in HCV-positive HCC were mainly involved in regulating metabolism (such as energy pathways, nucleic acid metabolism and amino acid and derivative metabolism) and cell proliferation related biological processes (including cell cycle, DNA repair, and DNA replication). Of note, emerging studies have indicated the important roles in HCC progression [28, 29]. However, very few reports have focused on exploring the roles of lncRNAs in HCC metabolism regulation. Our study for the first time provides useful information for exploring the effect of lncRNA on HCC metabolism.
The roles of most lncRNAs in HCC have remained unclear. In the present study, we identified lncRNAs (LOC341056, CCT6P1, PTTG3P, LOC643387, LOC100133920, C3P1 and C22orf45) as key lncRNAs. Furthermore, we constructed key lncRNA mediated mRNA-bp networks to reveal the potential roles of these key lncRNAs. Of these lncRNAs, PTTG3P has been reported to promote gastric tumor cell proliferation and invasion. However, the other lncRNAs were never reported in human diseases. According to bioinformatics analysis, we found that LOC341056, CCT6P1, PTTG3P, LOC643387, LOC100133920 were mainly involved in regulating metabolism and cell proliferation related processes, and C3P1 and C22orf45 were associated with immune response and energy pathways. Moreover, we performed Kaplan-Meier analysis to evaluate the prognostic value of these key lncRNAs. Higher expression levels of LOC643387, PTTG3P, LOC341056, CCT6P1 and lower C3P1 and C22orf45 expression were associated with shorter survival time in the TCGA dataset. These results suggested that these lncRNAs in HCV-positive HCC development could serve as biomarkers for hepatocellular carcinoma prognosis.
In conclusion, our study represents a comprehensive analysis of differentially expressed lncRNAs in HCV-positive HCC for the first time by analyzing the public dataset GSE17856. We identified 256 upregulated lncRNAs and 198 downregulated lncRNAs in HCV-positive HCC compared to the normal liver tissues. Co-expression network and GO analysis showed that these lncRNAs were involved in regulating metabolism, energy pathways, proliferation and immune response. Seven lncRNAs (LOC341056, CCT6P1, PTTG3P, LOC643387, LOC100133920, C3P1 and C22orf45) were identified as key lncRNAs and co-expressed with more than 100 DEGs in HCV-positive HCC. Kaplan-Meier analysis showed that higher expression levels of LOC643387, PTTG3P, LOC341056, CCT6P1 and lower expression levels of C3P1 and C22orf45 were associated with shorter survival time in the TCGA dataset. We believe that this study can provide novel potential therapeutic and prognostic targets for HCV-positive HCC.