Introduction
Esophageal carcinoma represents the sixth major factor responsible for malignancy-related deaths around the world [1, 2]. Esophageal cancer includes the adenocarcinoma (EAC) and squamous cell carcinoma (ESCC) types. The ESCC comprises almost 9 out of 10 cases in China. Although important advances exist in multimodal therapy of ESCC, prognosis remains poor, with a 5-year survival rate less than 15% [3]. Therefore, identifying reliable diagnostic and prognostic biomarkers of ESCC is critical, as well as elucidating molecular mechanisms participating in esophageal carcinogenesis [4, 5].
DEK was initially discovered as one of the parts of the DEK-nucleoporin 214 (DEK-NUP214, also known as DEK-CAN) fusion gene arising from a (6;9) translocation in a subtype of patients with acute myeloid leukemia (AML) [6]. Apart from being a fusion protein in AML, the DEK gene encodes a nuclear phosphoprotein that has been measured in many solid human tumors, such as hepatocellular carcinoma (HCC), lung cancer, bladder cancer, ovarian cancer and colorectal carcinoma [7–11]. Accumulating reports have indicated that DEK is involved in multiple cell metabolic processes, such as DNA damage repair, mRNA splicing, modifying chromatin structure and transcription regulation [12–17]. Several studies have also demonstrated that elevated DEK levels might accelerate tumorigenesis and neoplastic progression by means of intervention with cell proliferation, apoptosis, senescence, and differentiation [8, 18–22]. Zhou et al. reported that up-regulating DEK expression levels in A549 lung cancer cell lines could significantly promote cell proliferation, while restricting cell proliferation in DEK-depleted cells [8]. Liu et al. demonstrated that the silencing of DEK had effects on the growth of CaSki cells with blocking the cell cycle in the G0/G1 phase, inhibiting cell proliferation, increasing cell apoptosis and inducing cell senescence [20]. Wise-Draper et al. reported that the expression level of DEK protein was particularly repressed during keratinocyte differentiation and DEK overexpression in organotypic raft cultures was important for hyperplasia induction [22].
A large number of studies indicate that DEK is over-expressed in various malignant tumors, associated with tumorigenesis and advances in tumor invasion [7, 23–27]. For instance, researchers performed a comprehensive analysis of DEK expression in different melanocytic tumors and found that DEK expression was largely positive in malignant lesions, particularly in metastatic cases, while there was negligible DEK expression in benign lesions [23]. Javier et al. reported that the DEK gene was overexpressed in colorectal cancer cell lines, while knock-down of DEK in DLD1 and SW620 cell lines reduced cell migration and increased irinotecan-induced apoptosis [24]. Our previous study also demonstrated that mRNA level of DEK in the majority of hepatocellular carcinoma (HCC) tissues was approximately 1.8 times that in matched normal hepatic tissues (0.707 vs. 0.391), while the percentage of positive DEK expression in HCC tissues was about 2.19 times that rate in normal tissues (87.3% vs. 40.0%) [7]. Other studies have also shown that DEK frequently exhibits an increased expression level in various malignant tumors [25–27]. To date, there is only one study investigated the association between DEK expression and ESCC, by employing a DEK transgenic mouse model, and it found that DEK overexpression is capable of promoting ESCC in vivo [28]. Therefore, we conducted a hospital-based study to investigate the association between DEK expression in ESCC tissue samples, clinicopathologic features and patient survival.
Material and methods
Clinical samples
A total of 120 routinely diagnosed ESCC patients who underwent surgical resection at the Zhongshan Hospital, Xiamen University in the period from June 2011 to May 2013 were assessed. All cases were pathologically confirmed to be ESCC for the first time. Patients were excluded if any of the following conditions were met: (1) presence of tumors other than ESCC; (2) received chemotherapy or radiotherapy; (3) inoperable patients; (4) refusal or inability to participate in the investigation because of poor health. Pathological and demographical indexes, i.e. age, gender, tumor size, tumor-node-metastasis (TNM) stage, degree of tissue differentiation and lymph node metastasis (LNM), were obtained from medical records. Disease-free survival (DFS) and 5-year survival data were collected after follow-up. Our research has been approved by the Medical Ethical Committee of the Zhongshan Hospital, Xiamen University (Ethical Approval No. XMZSYYKY-2013-045) after reviewing the research protocol and a formal hearing. We ensured that written informed consent from patients was obtained before using samples in this study.
Immunohistochemical assessment
Immunohistochemical (IHC) assessment was completed with a DAKO LSAB kit (DAKO A/S, Glostrup, Denmark). 4 μm sections were submitted to deparaffinization in xylene, and rehydration in decreasing concentrations of ethanol (100%, 95%, 80% and 70%, respectively). Secondly, slides were incubated with 3% H2O2 in methanol for 15 min at room temperature. Subsequently, antigen retrieval was carried out by boiling in 0.01 M sodium citrate buffer (pH 6.0) for 20 min in a microwave. Sections were incubated in 20% normal goat serum for 20 min and were then incubated with the DEK-specific primary antibody (1 : 50; BD Biosciences, USA) at 4°C overnight according to the instructions. After incubation with biotinylated secondary antibody (ambient conditions, 30 min), samples were treated with streptavidin-peroxidase complex in ambient conditions for 30 min. Between each above-mentioned step, slides were washed with phosphate-buffered saline (PBS) for 5 min. Then, 3,3′-diaminobenzidine and hematoxylin were employed for development and counterstaining, respectively. Furthermore, samples processed without anti-DEK antibodies were employed as a negative control.
The slides were scored separately by two pathologists unaware of clinicopathological records. Firstly, the entire esophageal tumor was analyzed, and 50 randomly selected fields of view were quantitated, with nuclear staining considered to be positive. Briefly, the following scoring system was adopted for IHC staining: ‘–’ (< 5% positive cells), ‘+’ (5–25% positive cells), ‘++’ (26–50% positive cells) and ‘+++’ (> 50% positive cells) [29, 30]. DEK overexpression was reflected by ‘++’ or ‘+++’ score. In survival analyses, DEK expression was classified as low expression (‘–’ and ‘+’) and high expression (‘++’ and ‘+++’) [11, 26, 31].
Statistical analysis
SPSS 18.0 was employed for statistical analyses. Associations of DEK levels with clinicopathological features were assessed by the χ2 or Fisher’s exact test. Disease-free and 5-year survival rates upon resection were determined by Kaplan-Meier curves and compared by the log rank test. The Cox proportional hazards regression model was employed for multivariate survival analysis of characteristics with statistical significance in univariate analysis. The statistical significance level was set at p < 0.05.
Results
DEK is highly expressed at the protein level in ESCC
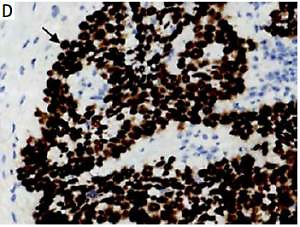
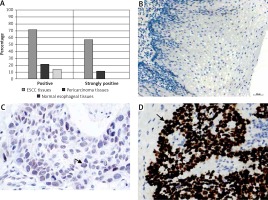
The DEK protein was exclusively found in the nucleus in ESCC tissues by IHC analysis (Figure 1 D). A rate of DEK expression of 71.7% (86/120) was obtained in esophageal cancer tissues, indicating a marked increase compared with the adjacent non-tumor (21.4%, 15/70) and normal esophageal mucosa (13.9%, 5/36) groups. Similarly, the rate of DEK overexpression in ESCC tissues (57.5%, 69/120) was markedly elevated in comparison with those of the adjacent noncancerous (11.4%, 8/70) and normal esophageal mucosa (0%, 0/36) groups (both p < 0.001) (Table I).
Figure 1
Immunochemical detection of DEK in esophageal tissues. A – Rates of expression and overexpression of DEK were considerably higher in ESCC in comparison with adjacent noncancerous or normal esophageal mucosal specimens. B – DEK protein not found in normal esophageal mucosa (original magnification, 200×). C – DEK protein found at low levels in pericarcinoma mucosa (original magnification, 400×). D – DEK protein overexpressed in ESCC tissue samples (original magnification, 400×)
Associations of clinicopathological parameters with DEK protein expression
The results showed a markedly increased rate of DEK expression in advanced-stage (III–IV) ESCC patients (49/55, 89.1%) compared with the early-stage (I–II) group (20/65, 30.8%) (p < 0.001). In addition, an elevated rate of DEK overexpression was found in poorly differentiated esophageal cancers (40/47, 85.1%) compared with moderately and well-differentiated cancers (29/73, 39.7%) (p < 0.001). However, DEK overexpression in esophageal cancer showed no associations with age, gender, tumor size or lymph node metastasis (p > 0.05) (Table II).
Table II
Associations between DEK expression and clinicopathological features of ESCC
Association of DEK expression with patient survival in ESCC determined by the Kaplan-Meier method
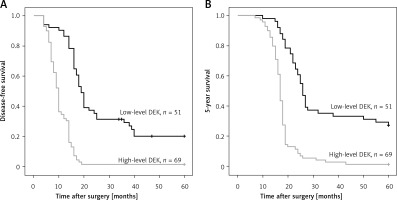
The ESCC cases (n = 120) were assessed by Kaplan-Meier survival curves. The results indicated that disease-free (log-rank p = 0.007) and 5-year (log-rank p = 0.008) survival rates were markedly elevated in ESCC patients with reduced DEK amounts compared with those highly expressing this protein (Figure 2).
DEK overexpression independently predicts prognosis in ESCC
Univariate analysis revealed that ESCC cases highly expressing DEK had markedly reduced disease-free and 5-year survival rates in comparison with patients lowly expressing DEK. In addition, TNM stage, differentiation degree, and lymph node metastasis (LNM) also showed significant associations with disease-free and 5-year survival rates. Multivariate analysis indicated that high TNM stage and LNM independently predicted poor prognosis in ESCC. It should be noted that high DEK expression was also confirmed to independently predict prognosis in ESCC (Table III).
Table III
Univariate and multivariate analyses by the Cox regression model for ESCC survival
| Characteristic | B | SE | Wald | HR | 95% CI | P-value | |
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| Univariate analysis: | |||||||
| Gender | 0.402 | 0.234 | 2.94 | 1.495 | 0.944 | 2.366 | 0.086 |
| Age | 0.365 | 0.238 | 2.364 | 1.441 | 0.904 | 2.296 | 0.124 |
| Tumor size | 0.458 | 0.235 | 3.808 | 1.582 | 0.998 | 2.507 | 0.051 |
| TNM stage | 1.224 | 0.214 | 32.596 | 3.399 | 2.233 | 5.174 | < 0.001** |
| Differentiation degree | 0.91 | 0.38 | 5.741 | 2.484 | 1.18 | 5.229 | 0.017* |
| Lymph node metastasis | 1.594 | 0.24 | 44.166 | 4.925 | 3.078 | 7.883 | < 0.001** |
| DEK | 1.375 | 0.22 | 39.068 | 3.957 | 2.571 | 6.091 | < 0.001** |
| Multivariate analysis: | |||||||
| TNM stage | 2.204 | 0.548 | 16.142 | 9.058 | 3.091 | 26.539 | < 0.001** |
| Differentiation degree | 0.352 | 0.518 | 0.464 | 1.423 | 0.516 | 3.923 | 0.496 |
| Lymph node metastasis | 1.121 | 0.543 | 4.26 | 3.069 | 1.058 | 8.899 | 0.039* |
| DEK | 1.416 | 0.422 | 11.272 | 4.121 | 1.803 | 9.42 | < 0.001** |
Discussion
Although significant associations of DEK with numerous cancers in humans have been reported, little is known about its effects in ESCC [28]. In this study, IHC analysis was performed to detect DEK protein expression in ESCC samples and para-carcinoma tissue specimens. As shown above (Table I, Figure 1), DEK was ubiquitously distributed in the nucleus of ESCC cells, with the rate of DEK expression (71.7%) markedly increased compared to para-carcinoma (21.4%) and normal esophageal tissues (13.9%, both p < 0.001). In agreement, the positive rate of DEK overexpression in ESCC tissues (57.5%) was markedly elevated compared with that in the para-carcinoma (11.4%) and normal esophageal samples (0%, both p < 0.001). The result was consistent with other studies in solid tumors which demonstrated that DEK was overexpressed in cancerous cells, such as HCC cells, lung cancer cells, bladder cancer cells and colorectal carcinoma cells [7–9, 11, 27, 29]. These findings provide valuable support for DEK as a potential diagnostic biomarker in solid tumors, including ESCC.
Next, associations of DEK overexpression with clinical and pathological indexes in ESCC were assessed. As shown above (Table II), the expression level of DEK correlated with TNM stage and the degree of tissue differentiation. The rate of DEK overexpression was significantly elevated in advanced-stage (III–IV) ESCC patients (89.1%) compared with early-stage (I–II) patients (30.8%, p < 0.001). Similarly, the rate of DEK overexpression was considerably increased in poorly differentiated esophageal cancer (85.1%) compared with moderately and well-differentiated cancers (39.7%, p < 0.001). Several studies have also reported that DEK overexpression was significantly related to clinical prognostic parameters [23, 31–33]. Liu et al. reported that DEK expression level is considerably higher in invasive ductal breast cancer compared with normal breast tissue and associated with proliferation (Ki-67 levels) and histological grade, indicating that DEK might potentially be an indicator for early detection and prognosis [32]. Sun et al. reported that DEK overexpression was strongly related to histological grade and TNM stage which indicated poor prognosis and recurrence in patients with pancreatic ductal adenocarcinoma (PDAC) [31]. Riveiro-Falkenbach et al. demonstrated that DEK expression was significantly positive in malignant lesions of melanoma, especially in metastatic cases, and correlated with histological features of aggressiveness [23]. In HPV16+ oropharyngeal squamous cell carcinoma, DEK overexpression was associated with advanced tumor stage and increased risk of death [33]. Our study indicated that elevated DEK expression might be a useful biomarker for evaluating ESCC progression and aggressiveness. In an in vivo study, Matrka et al. found that DEK overexpression could stimulate gross esophageal tumor development and cause a trend toward esophageal hyperplasia by employing a DEK transgenic mouse model [28].
In this study, ESCC cases with highly expressing DEK showed markedly reduced survival compared with those displaying low levels (Figure 2). Multivariate survival analysis confirmed DEK overexpression as an independent predictive factor of poor survival in ESCC, alongside TNM stage and lymph node metastasis (Table III). The association of DEK overexpression with disease-free survival or survival rates was consistent with other studies which reported that increased DEK expression was clearly correlated with poor prognosis in other solid tumors [7, 31, 32]. Sun et al. demonstrated that patients with high DEK expression had a lower overall survival rate than those with low DEK expression [31]. Liu et al. found that DEK protein levels were remarkably reduced in breast carcinoma cases with < 3-year disease-free survival (DFS) compared with those showing ≥ 3-year DFS [32]. Our previous study reported that DEK overexpression was significantly associated with portal vein invasion, tumor size and poor prognosis in HCC patients, showing that patients with high DEK expression had a shorter overall survival time than those with low expression [7].
In conclusion, based on the results obtained in the Cox regression model, DEK has been identified as a factor that is associated with reduced survival in both univariate analysis and multivariate analysis. Therefore, we can conclude that DEK expression is positively correlated with reduced survival in ESCC patients. Therefore, applying IHC in cancer tissues collected from ESCC patients to detect DEK expression would be helpful to evaluate the outcome after surgery. Our study has confirmed that DEK has potential to be an independent biomarker in predicting prognosis of ESCC patients. There are several limitations in the present study, which need to be stated. First of all, the sample size was limited. Secondly, the study was hospital-based, which may introduce selection bias of study subjects. Thirdly, we did not employ an animal model to investigate the impact of DEK on ESCC prognosis. Future investigations should be conducted to expand the sample volume among multi-center studies and test in vivo for understanding the straightforward role and mechanisms of DEK in ESCC malignancy.