Introduction
Uterine carcinosarcoma (CS) is a rare, biphasic tumour composed of high-grade carcinoma and sarcoma components, which account for less than 5% of all uterine tumours, while endometrial carcinoma (EC) represents approximately 95% of uterine tumours. The carcinomatous component is most often high-grade endometrioid or serous carcinomas, and uncommonly clear cell carcinoma [1].
The mesenchymal component is most commonly high-grade sarcoma not otherwise specified, but heterologous elements include rhabdomyosarcoma, chondrosarcoma, and osteosarcoma [1]. CS shows highly aggressive behaviour; at presentation, approximately 60% of patients with CS have advanced disease with extrauterine spread [2]. The prognosis of CS is also poor, with an overall 5-year survival rate of approximately 30% (stage I- II: 59%; stage III: 25%; stage IV: 9%) [3,4], in contrast to 90% in low-grade EC [5] and over 80% in stage I high grade EC [6]. Serous histology and hete-rologous rhabdomyoblastic differentiation have been reported to be significantly associated with worse survival [1].
Because of sampling and biopsy errors due to the fact that carcinosarcoma contains various components, the preoperative radiologic diagnosis of CS is clinically important. This dia-gnosis guides optimal surgical planning and adjuvant therapy, including novel targeted immunotherapy [7]. According to previous studies investigating MRI findings of uterine CS, large exophytic (12-75%), heterogeneous endo-metrial masses both on T2-weighted imaging (T2WI) 71-82%) and T1-weighted imaging (T1WI) (18-50%) have been reported to be suggestive of CS [8-10]. In dynamic studies, CSs typically show areas of early and persistent marked enhancement (50-100%) [8-10]. However, despite significant differences in prognosis between CS and EC, MRI findings of CS are far from sufficient to distinguish them from EC, including the report by Bharwani et al. showing that 88% of CS were indistinguishable from EC [8].
Convolutional neural network (CNN) is a class of deep learning models that combine imaging filters with artificial neural networks through a series of successive linear and nonlinear layers. It is considered a promising tool for diagnostic imaging, and several CNN models for diagnostic imaging have been constructed [11-14]. The modalities studied include radiography, US, CT, and MRI. However, to the best of our knowledge, no study has built a deep learning model for diagnosing CSs using MRI; therefore, its clinical usefulness requires validation.
Here, we present CNN models for differentiating CSs from ECs on MRI, including T2WI, apparent diffusion coefficient of water (ADC) map, and contrast enhanced-T1WI (CE-T1WI), and compare their diagnostic performance with interpretations by experienced radiologists.
Material and methods
Patients
This retrospective study was approved by the institutional review board of University of Tsukuba Hospital, and the requirement for written informed consent was waived (Approval number: R02-054).
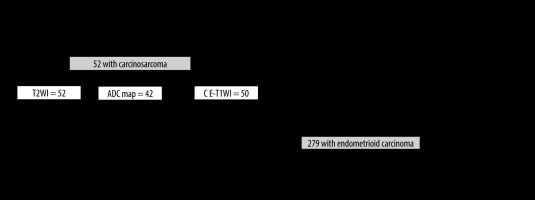
The inclusion criteria for the CS group were as follows: (a) pathologically proven CS by total hysterectomyand (b) a pelvic MRI in the oblique axial direction perpendicular to the long axis of the uterine body, performed at our hospital between January 2005 and December 2020. Note: Because CS is a rare tumour, cases of CS without diffusion-weighted imaging (DWI) or CE-T1WI were included. The inclusion criteria for the EC group were as follows: (a) pathologically proven EC by total hysterectomy and (b) a pelvic MRI including T2WI, DWI, and CE-T1WI in the oblique axial direction perpendicular to the long axis of the uterine body, performed at our hospital between January 2015 and December 2020. The exclusion criteria were as follows: (a) history of surgery other than caesarean section, chemotherapy, or radiation therapy of the uterus and (b) concomitant pregnancy. A flowchart for the patient selection process is shown in Figure 1.
MRI was performed using 3 T or 1.5 T equipment (Ingenia®, and Achieva®; Philips Medical Systems, Amsterdam, Netherlands). The imaging protocols for CS and EC are the same and included oblique axial T2WI, DWI with a b value of 0, 1000, and CE-T1WI with gadopentetate dimeglumine 5 mmol (Magnevist® or Gadovist®; Bayer, Wuppertal, Germany). The bolus intravenous contrast injection rate was 4 ml (2 mmol)/s (in case of Gadovist, diluted with saline solution and injected at 4 ml/s). Further details of these parameters are provided in Table 1.
Table 1
Magnetic resonance imaging acquisition parameters
[i] CE-T1WI – contrast-enhanced fat-saturated T1-weighted imaging, DWI – diffusion-weighted imaging, EPI – echo planar imaging, FA – flip angle, FOV – field of view, GRE SPIR – gradient echo spectral pre-saturation with inversion recovery, T2WI – T2-weighted imaging, TE – echo time, TR – repetition time, TSE – turbo-spin echo
Dataset
In order to detect the most suitable sequence for CNN to differentiate CS from EC, each sequence alone was targeted for this verification.
To create a dataset, only the slices in which the tumour was visualised were extracted based on the consensus of the 2 radiologists (T.S., A.U.) with reference to the pathology report. The same cross-section was extracted for all the sequences. A total of 331 patients were randomly assigned to the training and testing groups with the same ratio of CS to EC and a training-to-test ratio of approximately 5 : 1.
In the training phase of the CS group, 572 images of T2WI from 42 patients, 488 images of ADC map from 33 patients, and 539 images of CE-T1WI from 40 patients were used. In the training phase of the EC group, 1621 images of each sequence from 223 patients were used.
In the test phase, only 1 central image of the stacks was automatically extracted for each patient, and 66 images (10 images from 10 patients with CS and 56 images of 56 patients with EC) were used on T2WI and CE-T1WI, and 65 images (9 images from 9 patients with CS and 56 images from 56 patients with EC) were used on the ADC map.
In this study, the device used was unable to handle Digital Imaging and Communications in Medicine (DICOM) images, so the DICOM images were converted to Joint Photographic Experts Group (JPEG) images using the viewing software Centricity Universal Viewer (GE Healthcare, Chicago, IL, USA). Subsequently, the margins containing the patient information were automatically cropped and resized to 240 × 240 pixels using XnConvert (Gougelet Pierre-Emmanuel, Reims, France).
Deep learning with convolutional neural networks
Deep learning was performed on a deep station entry (UEI, Tokyo, Japan) with a GeForce RTX 2080Ti graphics processing unit (NVIDIA, Delaware, CA, USA), a Core i7-8700 central processing unit (Intel, Santa Clara, CA, USA), and deep-learning software Deep Analyzer (GHELIA, Tokyo, Japan) [15,16].
The conditions optimized based on the ablation and comparative studies of the previous research are as follows: CNN with Xception architecture [17] was used for deep learning. Xception is a novel architecture that has a similar parameter count as Inception V3, and inception modules have been replaced with depthwise separable convolutions. Therefore, the performance gains are not due to increased capacity but rather to more efficient use of model parameters. ImageNet [18] was used for pre-training. Optimiser algorithm = Adam (learning rate = 0.0001, β1 = 0.9, β2 = 0.999, eps = le-7, decay = 0, AMSGrad = false). The batch size was automatically selected by the Deep Analyzer to fit into the graphics processing unit (GPU) memory. Additionally, the horizontal flip, rotation (±4.5°), shearing (0.05), and zooming (0.05) were also automatically used as data augmentation techniques. The numbers of epochs used for the training were 50, 100, and 200. The validation/training ratio (validation ratio) was set at 0.1 or 0.2.
Radiologist interpretation
Three experienced radiologists (K.M., S.H., and M.S.) with 26, 12, and 8 years of experience in interpreting pelvic MRI independently reviewed the test images in a random order. Each image was evaluated by assigning confidence levels in the differentiation of CS and EC using a 6-point scale (0, definitely CS; 0.2, probably CS; 0.4, possibly CS; 0.6, possibly EC; 0.8, probably EC; and 1.0, definitely EC). They were blinded to pathological and clinical findings. A duration of a week or more was observed between each sequence interpretation. Prior to interpretation, several cases of CS and EC were presented, and the typical imaging findings of CS (large exophytic, heterogeneous endometrial masses with marked enhancement) were reconfirmed.
Statistical analysis
The age and tumour stage for each group were compared using the Mann-Whitney U test and χ2 test.
The test dataset was used to calculate the sensitivity, specificity, and accuracy of the CS diagnoses. In radiologists, a value of 0-0.4 was treated as CS and 0.6-1.0 was treated as EC. In CNN, the classification into CS and EC groups was by output as a continuous number from 0 to 1, where 0-0.49 was considered as CS and 0.50-1.00 was considered as EC. Receiver operating characteristic (ROC) analysis was performed to assess the diagnostic performance [19]. For statistics, 95% confidence intervals (CIs) and significance difference were estimated. In addition, the inter-observer agreement for the 2 choices of CS or EC was assessed using kappa (κ) statistics. The κ-statistic interpreted the agreement as follows: less than 0, no; 0-0.20, slight; 0.21-0.40, fair; 0.41-0.60, moderate; 0.61-0.80, substantial; and 0.81-1.00, almost perfect [20].
All statistical analyses were performed using SPSS software (SPSS Statistics 27.0; IBM, New York, NY, USA). The statistical significance was set at p < 0.05.
Results
A total of 331 women (mean age 59 years; age range 30-91 years) were evaluated across the datasets. Table 2 shows the patients’ characteristics, clinical stages, and image slices of CS and EC lesions. There was no significant difference between the CS and EC groups and training and testing data regarding patient age and tumour stage. In the CS group, 7 (train 5, test 2) patients were scanned at 1.5 T. In the EC group, 13 (train 10, test 3) patients were scanned at 1.5 T. The other patients were scanned at 3 T.
Table 2
Characteristics of patients and lesions
For the selection of the validation ratio and epochs among the models, a model with a validation ratio of 0.2 and 100 epochs was adopted for T2WI (Figure 2), a model with a validation ratio of 0.1 and 100 epochs was adopted for the ADC map, while a model with a validation ratio of 0.2 and 200 epochs was adopted for CE-T1WI, owing to their high diagnostic performance.
Figure 2
Accuracy and loss of the training data (T2-weighted imaging with validation ratio 0.2, epochs 100)
Table 3 presents the results of the interpretation. Table 4 lists the diagnostic performances of the CNN models and the 3 radiologists in diagnosing CS. The ROC curves comparing the performance of the CNN models and the radiologists are shown in Figure 3; the sensitivity, specificity, accuracy, and AUC of the CNN model of each sequence in diagnosing CS were comparable to those of Reader 1 and superior to those of the other 2 readers. The CNN models and Reader 1 were able to differentiate between CS and EC, with significant differences in all sequences. And the AUC was significantly higher on T2WI and CE-T1WI for the CNN model than for Readers 2 and 3. The CNN model showed the highest diagnostic performance on the T2WI with an AUC of 0.94 when comparing the sequences and interpreters.
Table 3
Interpretation results of the CNN model and the radiologists
| CNN | Reader 1 | Reader 2 | Reader 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CS | EC | CS | EC | CS | EC | CS | EC | ||
| T2WI | CS | 7 | 3 | 7 | 3 | 3 | 7 | 6 | 4 |
| EC | 4 | 52 | 9 | 47 | 13 | 43 | 31 | 25 | |
| ADC map | CS | 4 | 5 | 7 | 2 | 3 | 6 | 5 | 4 |
| EC | 6 | 50 | 7 | 49 | 13 | 43 | 32 | 24 | |
| CE-T1WI | CS | 7 | 3 | 7 | 3 | 3 | 7 | 6 | 4 |
| EC | 4 | 52 | 5 | 51 | 13 | 43 | 31 | 25 | |
Table 4
Sensitivity, specificity, and AUC of the CNN model and the radiologists in diagnosing CS
[i] ADC – apparent diffusion coefficient, AUC – area under the receiver operating characteristic curve, CE-T1WI – contrast-enhanced fat-saturated T1-weighted imaging, CI – confidence interval, CNN – convolutional neural network, T2WI – T2-weighted imaging. Data in parentheses are 95% confidence interval. *p < 0.05
Figure 3
Receiver operating characteristic (ROC) curves for the performance of the convolutional neural network (CNN) model compared to radiologists’ performance
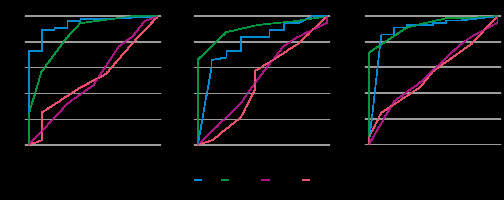
Table 5 shows the inter-observer agreement between the CNN models and the 3 radiologists. The CNN model and Reader 1 of T2WI and CE-T1WI and Reader 2 and 3 of all sequences showed a fair agreement. The other studies showed no agreement.
Table 5
Inter-observer agreement between the CNN model and the radio-logists
| Reader 1 | Reader 2 | Reader 3 | ||
|---|---|---|---|---|
| T2WI | CNN | 0.59 | 0.25 | 0.05 |
| Reader 2 | 0.16 | – | – | |
| Reader 3 | 0.01 | 0.40 | – | |
| ADC map | CNN | 0.09 | 0.05 | 0.07 |
| Reader 2 | 0.04 | – | – | |
| Reader 3 | 0.01 | 0.40 | – | |
| CE-T1WI | CNN | 0.53 | 0.03 | 0.05 |
| Reader 2 | 0.10 | – | – | |
| Reader 3 | 0.13 | 0.40 | – | |
Figures 4-6 show the test images of this study with interpretations of the CNN model and the radiologists, including the confidence of the CNN model and the radiologists. Figure 4 is a typical CS case, where the CNN model and Reader 1 diagnosed CS on T2WI. On the other hand, only the CNN model could not diagnose CS on the ADC map, and all but Reader 2 diagnosed CS on CE-T1WI. Figure 5 presents a case of CS of a huge tumour with internal necrosis, where all but Reader 2 diagnosed CS on T2WI and ADC map, and only the CNN model diagnosed CS on CE-T1WI. Figure 6 shows a case of EC with haemorrhage in the endometrial cavity, where the CNN model diagnosed EC only on CE-T1WI.
Figure 4
A 72-year-old woman with carcinosarcoma showing heterologous differentiation of stage IVB. T2WI: the CNN model = CS (confidence; CS = 100.0%), Reader 1 = CS (confidence; CS = 100.0%), Reader 2 = EC (confidence; CS = 0.0%), Reader 3 = EC (confidence; CS = 40.0%). ADC map: the CNN model = EC (confidence; CS = 0.2%), Reader 1 = CS (confidence; CS = 80.0%), Reader 2 = CS (confidence; CS= 60.0%), Reader 3 = CS (confidence; CS = 80.0%). CE-T1WI: the CNN model = CS (confidence; CS = 100.0%), Reader 1 = CS (confidence; CS = 100.0%), Reader 2 = EC (confidence; CS = 20.0%), Reader 3 = CS (confidence; CS = 100.0%). The tumour showed relatively heterogeneous hyperintensities both on T2WI and ADC map (arrow heads); additionally, some parts were strongly enhanced on CE-T1WI (arrow), which were typical appearances of CS. However, it seemed difficult to distinguish it from EC associated with endometrial polyp. It may also have been difficult to distinguish from adenomyosis with the ADC map alone
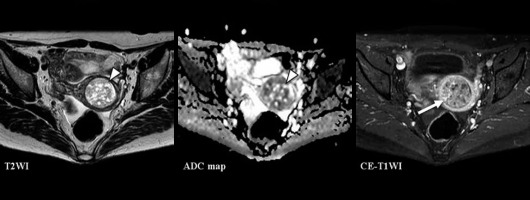
Figure 5
A 74-year-old woman with carcinosarcoma showing heterologous differentiation of stage IVB. T2WI: the CNN model = CS (confidence; CS = 100.0%), Reader 1 = CS (confidence; CS = 100.0%), Reader 2 = EC (confidence; CS = 40.0%), Reader 3 = CS (confidence; CS = 60.0%). ADC map: the CNN model = CS (confidence; CS = 69.8%), Reader 1 = CS (confidence; CS = 100.0%), Reader 2 = EC (confidence; CS = 40.0%), Reader 3 = CS (confidence; CS = 100.0%). CE-T1WI: the CNN model = CS (confidence; CS= 100.0%), Reader 1 = EC (confidence; CS = 40.0%), Reader 2 = EC (confidence; CS = 40.0%), Reader 3 = EC (confidence; CS = 40.0%). Huge tumour with exophytic growth was accompanied by extensive necrosis and showed heterogeneous signal intensities both on T2WI and ADC map (arrow heads). In CE-T1WI, the tumour enhancing effect was poorer than that in the myometrium (arrow), and no enhancing effect was observed in the necrotic area. It is presumed that all readers diagnosed EC because the tumour enhancing effect was poor on CE-T1WI
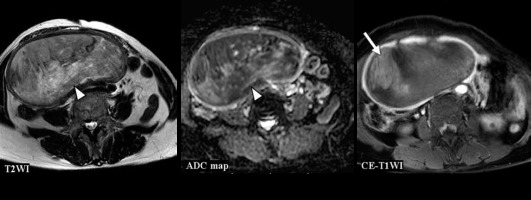
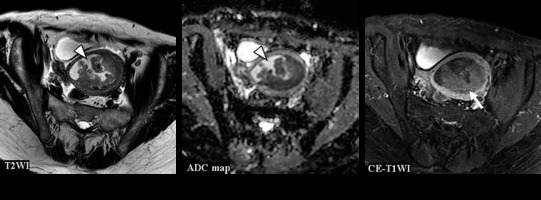
Figure 6
A 63-year-old woman with endometrioid carcinoma Grade 1 of stage IVB. T2WI: the CNN model = CS (confidence; CS= 100.0%), Reader 1 = CS (confidence; CS = 60.0%), Reader 2 = EC (confidence; CS= 0.0%), Reader 3 = CS (confidence; CS = 60.0%). ADC map: the CNN model = CS (confidence; CS = 96.6%), Reader 1 = EC (confidence; CS = 40.0%), Reader 2 = EC (confidence; CS = 40.0%), Reader 3 = CS (confidence; CS = 100.0%). CE-T1WI: the CNN model = EC (confidence; CS = 0.1%), Reader 1 = CS (confidence; CS = 60.0%), Reader 2 = EC (confidence; CS = 40.0%), Reader 3 = CS (confidence; CS = 80.0%). Haemorrhage in the endometrial cavity resulted in heterogeneity (arrow heads), which may have made interpretation difficult. However, it may have been possible to make a correct diagnosis because the tumour showed a relatively homogeneous poor enhancement on CE-T1WI (arrow)
Discussion
This study presented CNN models to distinguish between CS and EC using several MRI sequences, which demonstrated almost equal diagnostic performance compared to the most experienced radiologist, Reader 1, and better diagnostic performance compared to Readers 2 and 3.
Few studies have used deep learning in the field of uterine tumour MRI. Urushibara et al. developed a CNN that can differentiate between cervical cancer and non-cancerous lesions on single sagittal T2WI and showed high diagnostic performance [16]. A study by Chen et al. [21] and Don et al. [22] used T2WI, T2WI, and CE-T1WI, respectively, to evaluate the myometrial infiltration of ECs using CNN and showed comparable diagnostic performance compared to radiologists. To the best of our knowledge, this is the first study on the diagnosis of CS in deep learning using MRI.
Large exophytic heterogeneous endometrial masses have been reported to be suggestive of CS [23]. Haemorrhagic necrosis is often observed in large masses, leading to heterogeneity on T2WI and T1WI [8-10]. In dynamic studies, CS typically shows areas of early and persistent marked enhancement, similar to that of the myometrium, and/or a gradual and delayed strong enhancement, contrary to EC with consistently poor enhancement [8-10]. According to previous studies of Bharwani et al. with 51 CS cases [8], Tanaka et al. with 17 CS cases [9], and Ohguri et al. with 4 CS cases [10], large exophytic appearance was seen in 12% [8], 100% [9], and 75% [10], heterogeneity on T2WI was seen in 82% [8], 71% [9], and 75% [10], heterogeneity on T1WI was seen in 33% [8], 18% [9], and 50% [10], heterogeneity on CE-T1WIs was seen in 58% [8], 50% [9], and 100% [10], and delayed strong enhancement was seen in 50% [8], 81% [9], and 100% [10], repsectively. Garza et al. generated time-intensity curves for tumour and surrounding myometrial regions of interest in 37 patients with CS and 42 patients with EC, measuring the positive enhancement integral (PEI), maximum slope of increase (MSI), and signal enhancement ratio (SER). The threshold PEI ratio ≥ 0.67 predicted CS with 76% sensitivity, 84% specificity, and 0.83 AUC, and the threshold SER ≤ 125 predicted CS with 90% sensitivity, 50% specificity, and 0.72 AUC; however, tumour MSI did not significantly differ between the disease groups [24]. It has been reported that there was no significant difference between the mean and minimum ADC values of CS and EC, although the mean ADC of CS was significantly higher than that of higher grade (Grade 2 and 3) EC [25], and CS demonstrated heterogenous signal intensities on DWI and ADC map reflecting complicated tissue components [26]. In summary, heterogeneous signal intensities indicating haemorrhagic necrosis, and strongly enhanced areas are predictors of uterine CSs on MRI [10]. However, the dia-gnostic performance of imaging features is not perfect. Especially when the tumour is small or lacks these typical diagnostic imaging features, preoperative diagnosis is not always possible and there is an oportunity for the use of deep learning to assist in interpretation.
The MRI findings of CS are not sufficient to differen-tiate them from EC. In addition, our CNN models consisted of only 42 CS cases, and although the images used were not cropped images of the uterus only, our CNN models showed diagnostic performances equivalent to those of experienced radiologists. The CNN model showed the highest diagnostic performance with T2WI. In contrast, the low diagnostic performance on ADC map compared to the other sequences was inconsistent with previous reports [27], and it was presumed to result from the relatively small number of training ADC maps.
The inter-observer agreement between the readers, not only between the CNN model and the readers, was gene-rally low. This indicates that the difference in the imaging findings between CS and EC was not standardised or generalised even among experienced radiologists, and CS and EC could not be clearly classified.
Our study had several limitations. First, the number of CSs was small; however, this was the largest number of MRI studies of CSs to date. Second, each sequence was evaluated individually, which was quite different from the clinical practice wherein all sequences were referenced and comprehensively diagnosed. However, our unpublished data indicated that this case size could not be expected to improve interpretation by using a combined image set. Also, unlike radiologists, one of the CNNs’ strengths may be the ability to demonstrate high diagnostic performance with only one image of one type of sequence. In addition, even when diagnosing with combined sequences by CNN, weighting each sequence will be required. Third, due to the rarity of CS, the period of adoption for CS and EC was different. Even if there is a large amount of data on one side, good results cannot be obtained. In addition, the number of images of each sequence available in CS was also different, which may have affected the results. Finally, we did not examine T1WI and dynamic studies, to avoid study complexity. Although T1WI is useful for assessing haemorrhage, it was not included in this study because the scan protocol did not include oblique axial T1WI. Moreover, we considered that the same information is included in CE-T1WI. Dynamic studies have been reported to help distinguish between CS and EC; however, we believe that delayed isolation and hyperenhancement in the late contrast phase are of utmost importance to distinguish between the 2, and that dynamic study is not essential.
The following can be considered as future improvements: For rare tumours, such as CS, it is necessary to accumulate cases at multiple centres and increase the number of images. As mentioned in the study limitations, deep-learning models trained using a combination of several sequences may show superior diagnostic performance to the current models, as reported for the CNN model of prostate cancer using fused ADC and T2WI [28]; however, additional training images may be required to achieve a higher diagnostic performance with combination images. Evaluating series images as well as learning with DICOM data can also improve diagnostic performance, and greater versatility can be achieved by using images taken with other MRI instruments. In this study, we used ImageNet [18], which comprised natural images as pre-trained data, and the diagnostic performance could be further improved if transfer learning using medical image training data [29] was used.
Conclusions
CS is much less prevalent than EC and challenging to distinguish from EC; however, under the limited conditions of this study, deep learning provided comparable or better diagnostic performance to experienced radiologists when distinguishing between CS and EC on MRI. Although further validation is needed, deep learning has the potential to help with highly specialized MRI image interpretation.