Introduction
Rheumatoid arthritis (RA) is a kind of autoimmune disease with primarily symmetrical polyarthritis. Inflammatory cell infiltration, synovial tissue dysplasia and progressive destruction of articular cartilage and bone are the main features of this disease. RA synovial fibroblast-like cells (RASF) that originate from the synovial tissue lining display an activated RA phenotype and function, including abnormal cell proliferation, survival, growth and migration, which are regulated by members of cell cycle regulator families [1].
Currently, proteins such as cyclins, cyclin-dependent kinases or E2F transcription factor (E2F) are being studied in the context of cell proliferation and survival, cell signaling and cell cycle regulation [2]. Members of the E2F family have a common helix-loop-helix structure, usually form heterodimers, and bind the promoter of target genes mediating the cell cycle [3]. The E2F family includes nine proteins encoded by eight genes, namely, E2F1, E2F2, E2F3a, E2F3b, E2F4, E2F5, E2F6, E2F7 and E2F8 [4]. As an important factor in the regulation of cell mitotic events, the nuclear transcription factor E2F2 regulates cell division and cell cycle-related genes in lymphocytes [5-7]. The E2F2 protein is transported from the cytoplasm into the nucleus, where it binds to a gene promoter to stimulate the transcription of target genes and plays a major role in the G1/S transition in the mammalian and plant cell cycle [8]. Previously, using Illumina Human HT-12 v4 Expression BeadChip, we screened novel RA-specific gene expression by simultaneously comparing the expression profiles of synovial tissues from patients with RA, osteoarthritis (OA) and ankylosing spondylitis (AS). By comparing the global gene profiles in synovial tissues from the three diseases, we finally identified a specific increase in E2F transcription factor 2 (E2F2) in RA synovial tissues. Our immunohistochemical analysis also revealed significant E2F2 expression in synovial membranes from RA patients. In addition, we detected reduced interleukin (IL)-1α and IL-1β expression levels in the culture medium of RASF transfected with anti-E2F2 siRNA. Additionally, our observations demonstrated that synovial fibroblasts, the main cells in the synovium, have high expression of E2F2. This study suggests that the uniquely increased expression of E2F2 in RA synovial tissues may activate the disease process [9]. However, the pathogenic mechanism and the effect of E2F2 in RA are not well known. The present study aimed to investigate the pathogenic function and downstream signaling pathway of E2F2 in RA.
Material and methods
Rheumatoid arthritis synovial tissue collection
Human synovial tissues were collected from patients with RA (n = 10, 7 females, 28-78 years old, mean age of 56) during knee joint arthroscopic synovectomy. The diagnosis conformed to the revised criteria of the American College of Rheumatology. Patients with RA were medicated with non-steroidal anti-inflammatory drugs, which helped reduce the pain and swelling of the joints and decreased stiffness. Patients were enrolled between May 2015 and October 2016 at Shandong Provincial Qianfoshan Hospital. All patients signed written informed consent for the study. The study protocol was approved by the Medical Ethical Committee of Shandong Provincial Qianfoshan Hospital at Jinan, China (2015031).
Preparation and culture of RASF
Synovial tissues were finely minced and incubated with Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA) containing 1 mg/ml collagenase type II (Sigma-Aldrich, St. Louis, MO, USA) for 6 h at 37oC with 5% CO2. An equal volume of phosphate-buffered saline (PBS) containing 0.25% trypsin (Solabio, Beijing, China) was added to the culture, and the incubation continued for an additional 1 h. The cells were washed and re-suspended in DMEM supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA) and 1% penicillin/streptomycin (Solabio, Beijing, China). Cells were cultured overnight, and floating non-adherent cells were removed. In this study, RASF were passaged through five generations to the logarithmic growth phase.
Silencing E2F2 expression in RASF using siRNA
Guangzhou Ruibo Company (Guangzhou, China) designed and produced the anti-E2F2 siRNA. The target sequence of the anti-E2F2 siRNA was CTGCAGATATATCTCAAGA. SiRNA transfection was performed using HiPerFect transfection reagent (Qiagen) according to the manufacturer’s instructions. A parallel experiment was conducted with the Allstars siRNA provided with the kit. This siRNA does not suppress the expression of any genes and is normally used as a negative control. RASF were suspended in the appropriate culture medium, added to each well of six-well culture plates and incubated under normal growth conditions (37oC and 5% CO2) before transfection. When cell confluence reached 70% in each well, anti-E2F2 siRNA or Allstars siRNA and HiPerFect transfection reagent (Qiagen, Germany) were added to serum-free culture medium. E2F2 expression knockdown was verified by real-time polymerase chain reaction (PCR) and Western blot analysis.
RNA extraction and real-time PCR
Total RNA was extracted from cultured RASF using TRIzol reagent (Sigma, USA) according to the manufacturer’s protocol. cDNA was synthesized using an RNA PCR kit (Toyobo, Japan). Real-time PCR was conducted using a ViiA7Dx instrument (ABI, USA) according to the manufacturer’s instructions. The relative expression levels of the target genes were normalized to the relative GAPDH mRNA levels. The experiment was performed in triplicate, and the PCR products were verified using melting curve analysis. The relative mRNA expression level was calculated using the Ct method with the following formula: 2–ΔΔCt = 2–ΔCt (target gene) – ΔCt (GAPDH). The mRNA expression of the target genes was quantified with fold change by comparison with the GAPDH expression level. Shenggong (Shanghai, China) designed and provided the commercial primers. The primers designed for the targeted genes are shown in Table 1.
Table 1
PCR primer sequences
RT2 profiler PCR array
A human inflammatory RT2 Profiler PCR Array (Qiagen) was used to screen inflammatory genes that were affected by E2F2 expression. This PCR array profiled the expression of 84 different key genes that are commonly involved in the dysregulation of signal transduction and other normal biological processes during inflammation and autoimmune reactions. PCR array analysis was conducted using a ViiA7 DX according to the manufacturer’s instructions. RASF from patients (n = 10, including 7 women) were transfected with anti-E2F2 siRNA, and the inhibition of E2F2 expression was verified using real-time PCR and Western blotting. Equal amounts of total RNA (100 ng from each sample) from 10 synovial samples were pooled to minimize individual differences. Total RNA was reversed-transcribed into cDNA using an RT2 First Strand Kit, as described above. Subsequently, cDNA was mixed with an appropriate amount of RT2 SYBR Green master mix. This mixture was aliquoted into the wells of the RT2 Profiler PCR array. PCR was performed under the following thermal cycling conditions: 95oC for 10 min, 40 cycles of 95oC for 15 s and 1 cycle of 60oC for 1 min. Another PCR array experiment was prepared by using total RNA extracted from the cells treated with the Allstars siRNA. The transcription levels of target genes were normalized to their corresponding expression levels in the Allstars siRNA-transfected cells. Data analysis was performed using the Web-based RT2 Profiler PCR Array Data Analysis software (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php, SA Biosciences). Fold changes in gene expression were calculated and expressed as log normalized ratios of the expression level in anti-E2F2 siRNA-transfected cells/controls according to the manufacturer’s instructions. Based on our experience using PCR arrays, at least a twofold change in gene expression was considered biologically significant in this study. The PCR array results were confirmed by real-time PCR using RNA extracts from 10 anti-E2F2 siRNA-transfected RASF samples.
E2F2 overexpression in RASF via transfection of E2F2-expressing plasmids
Shanghai Boshang Company (China) constructed E2F2-expressing plasmids. The full E2F2 coding sequence is 1314-bp long and codes 431 amino acid residues. The full coding sequence was amplified from cDNA, and the cDNA had been reverse-transcribed from total RNA using the following specific primers: E2F2-HindIII-Fex (GGCAAGCTTCGATGCTG CAAGGGCCCCGGGCC) and E2F2-BamHI-Rex (TGCGGATCCTCAATTAATCAACAGGTCCCCAAGGTC). The PCR product was then sequenced and inserted into pEGFP-C1 vectors (Clontech) with HindIII and BamHI clone sites. RASF from those ten patients were suspended in appropriate culture medium, added to each well of six-well plates and incubated under normal growth conditions (37oC with 5% CO2). When the cell density reached 70% in each well, E2F2-expressing plasmid DNA and HiPerFect transfection reagent (Genmute, USA) were mixed, and the cell incubation continued. The expression levels of targeted genes were determined by real-time PCR in the RASF samples transfected with E2F2-expressing plasmids.
Western blot analysis
RASF transfected with anti-E2F2 siRNA or E2F2-expressing plasmids were homogenized with cell lysis solution (Beyotime, China) and centrifuged at 12,000 × γ for 30 min at 4oC. The supernatant was collected, and protein concentrations were determined using a BCA protein assay kit (Beyotime, China). Total protein was separated on 10% SDS-PAGE gels, transferred to PVDF membranes and blocked in 5% milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) at pH 7.5. The membranes were incubated overnight at 4oC with rabbit polyclonal antibody against human E2F2 (ab138515, Abcam, UK), goat polyclonal antibody against human CCR4 (ab1669, Abcam, UK) or rabbit polyclonal antibody against human GAPDH (Good Here, China) at a 1 : 1000 dilution. Horseradish peroxidase-conjugated sheep anti-rabbit immunoglobulin G (IgG) secondary antibody or rabbit anti-goat IgG secondary antibody was incubated with the membrane for 1 h at room temperature in TBST. Complexes were visualized using Immobilon Western chemiluminescent HRP substrate (Millipore, USA). The expression level of E2F2 or CCR4 protein was quantified by normalizing GAPDH expression using Image J software (NIH, Bethesda, MD, USA) to compare their gray values.
Determining RASF apoptosis using flow cytometry
RASF from those ten patients were transfected with anti-E2F2 siRNA or E2F2-expressing plasmids. The transfected RASF were trypsinized and then centrifuged at 200 × γ for 5 minutes, followed by washing with PBS. After removing the supernatants, the cell density was adjusted to 1-5 × 106 cells/100 µl. Cell apoptosis was determined using an apoptosis kit (Neobioscience, Shenzhen, China) according to the manufacturer’s manual. Flow cytometry was conducted with a FACSAria instrument (BD Biosciences, Franklin Lakes, NJ, USA). Each test was performed in triplicate.
Determining the migration ability of RASF using transwell assay
A Transwell chamber (Corning Inc., Corning, NY, USA) was used to evaluate the migration ability of RASF from those ten patients. RASF from those ten patients were transfected with anti-E2F2 siRNA or E2F2-expressing plasmids. The transfected RASF were suspended in serum-free medium. Cell density was adjusted to 2 × 104 cells/ml, and 100 µl of cell suspension was added to the transwell chambers. Then, the medium containing 2.5% serum was added into the lower chamber. After incubating at 37oC for 24 h, the cells in the upper chamber were wiped with a cotton swab and fixed with methanol. After being stained with 0.1% crystal violet, the number of cells was counted under a microscope.
Tube-like structure formation assay with RASF
Matrigel (Corning Life Sciences) was added dropwise into 24-well culture plates and allowed to polymerize for 1 h at 37oC. RASF from those ten patients were transfected with anti-E2F2 siRNA or E2F2-expressing plasmids were seeded onto the 3D matrix in complete medium. The number of tube-like structures was counted using phase contrast microscopy (original magnification 10×). Twelve microscopic fields were randomly selected from each well, and the number of tube-like structures per field was counted.
Statistical analysis
All data were processed using SPSS Statistics 19.0 software. Multiple comparisons were conducted using ANOVA. T-tests were conducted to assess significant differences between two groups. P values less than 0.05 were considered significant. Data are presented as standard deviations.
Results
Investigating E2F2 pathogenic pathway in RASF
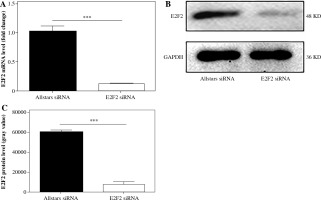
Ten RASF samples from patients were transfected with anti-E2F2 siRNA to inhibit E2F2 expression. The E2F2 mRNA expression level in RASF was examined using real-time PCR. E2F2 mRNA expression was significantly decreased in these 10 RASF samples compared with Allstars siRNA-transfected RASF samples (p = 0.0037). The E2F2 protein expression level in RASF was also examined using Western blot analysis. E2F2 protein level was significantly decreased in these RASF samples compared with Allstars siRNA-transfected RASF samples (p = 0.0075). The result is shown in Figure 1.
Fig. 1
E2F2 expression level in RASF with transfection of anti-E2F2 siRNA. A) E2F2 mRNA expression level in RASF was detected using real-time PCR. B) E2F2 protein expression was detected in RASF using Western blot analysis. C) E2F2 protein expression was normalized to GAPDH expression. ***p < 0.001
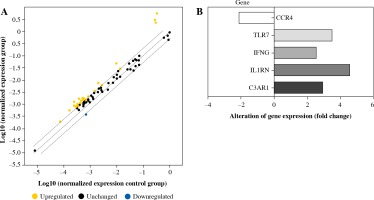
PCR array was used to examine the expression of 84 immunity and inflammation-related genes. Five genes had more than a twofold change in expression between E2F2-inhibited RASF and RASF controls transfected with Allstars siRNA. The expression of complement component 3a receptor 1 (C3AR1), interleukin 1 receptor antagonist (IL1RN), interferon γ (IFN-γ) and Toll-like receptor 7 (TLR7) was significantly up-regulated, and the expression of C-C chemokine receptor type (CCR4) was significantly down-regulated in RASF with inhibited E2F2 expression. The results are shown in Figure 2.
Fig. 2
PCR array analysis of the gene expression profile in RASF transfected with anti-E2F2 siRNA. A) Detection of relative gene expression levels in RASF using a human inflammatory PCR array. B) The PCR array result is depicted in one map
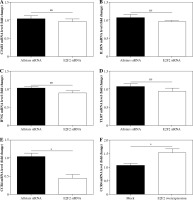
The genes screened by PCR array were verified using real-time PCR analysis with those 10 RASF samples with transfection of anti-E2F2 siRNA. Compared with the expression level in RASF transfected with Allstars siRNA, C3AR1 expression was up-regulated in 8 of the 10 samples but was down-regulated in 2 samples; CCR4 expression was down-regulated in 9 samples but was up-regulated in 1 sample; IFN-γ expression was down-regulated in 7 samples but was up-regulated in 3 samples; IL1RN expression was up-regulated in 7 samples but down-regulated in 3 samples; TLR7 expression was down-regulated in 7 samples but was up-regulated in 3 samples. After statistical analysis, CCR4 expression was finally determined to be significantly decreased (p = 0.0144). These results are shown in Figure 3A-E.
Fig. 3
mRNA expression of the screened genes by PCR array analysis in RASF. A) C3AR1, B) IL1RN, C) IFNG, D) TLR7, E) CCR4 mRNA expression level in RASF with transfection of anti-E2F2 siRNA or Allstars siRNA. F) CCR4 mRNA expression level in RASF with transfection of E2F2-expressing plasmids or the blank plasmids (mock). *p < 0.05, ns – no statistical significance
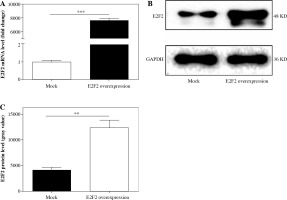
Those 10 RASF samples were also transfected with E2F2-expressing plasmids, and the E2F2 mRNA level was verified using real-time PCR. The E2F2 mRNA expression level was significantly increased in RASF transfected with E2F2-expressing plasmids in all 10 samples (p = 0.001). The E2F2 expression level in RASF was also examined using Western blot analysis. E2F2 protein level was significantly increased in the 10 RASF samples compared with the blank plasmids-transfected RASF samples (p = 0.0044). The above results are shown in Figure 4.
Fig. 4
E2F2 expression level in RASF with transfection of E2F2-expressing plasmids or blank plasmids (mock). A) E2F2 mRNA expression level in RASF was detected using real-time PCR. B) E2F2 protein expression was detected in RASF using Western blot analysis. C) E2F2 protein expression was normalized to GAPDH expression. **p < 0.01, ***p < 0.001
In these plasmids-transfected RASF samples, CCR4 mRNA expression was up-regulated in 9 samples but was down-regulated in 1 sample. After statistical analysis, the results indicated that CCR4 expression was significantly increased in the RASF samples with E2F2 overexpression (p = 0.0205). This result is shown in Figure 3F.
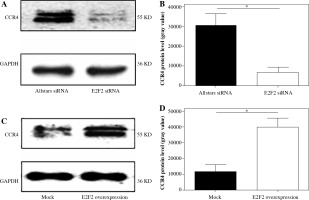
Western blot analysis was performed to examine CCR4 protein expression in the 10 RASF samples transfected with anti-E2F2 siRNA or E2F2-expressing plasmids. CCR4 protein expression was significantly decreased in the E2F2-silenced RASF compared with Allstars siRNA-transfected RASF (p = 0.0139). E2F2 protein expression was significantly increased in RASF transfected with E2F2 overexpression compared with RASF transfected with the blank plasmids (p = 0.0442). The results are shown in Figure 5.
Fig. 5
CCR4 protein expression in RASF using Western blot analysis. A) CCR4 protein expression in RASF transfected with anti-E2F2 siRNA. B) CCR4 expression was normalized to GAPDH expression in the siRNA-transfected RASF. C) CCR4 expression in RASF transfected with E2F2-expressing plasmids. D) CCR4 expression was normalized to GAPDH expression in the E2F2-expressing RASF. *p < 0.05
Investigating the effect of E2F2 expression on RASF
To determine the effect of E2F2 expression on RASF phenotype and activity, those 10 RASF were transfected with anti-E2F2 siRNA. Flow cytometry was performed to determine the effect of E2F2 expression on RASF apoptosis. The assay showed that early apoptosis was significantly increased in RASF following transfection of anti-E2F2 siRNA compared with the cells transfected with Allstars siRNA (p = 0.0435). This result is shown in Figure 6A. Those 10 RASF were also transfected with E2F2-expressing plasmids. Flow cytometry showed that early apoptosis was significantly decreased in RASF following transfection of the E2F2-expressing plasmids compared with the cells transfected with the blank plasmids (p = 0.025). This result is shown in Figure 6B.
Fig. 6
The effect of E2F2 expression on apoptosis of RASF using flow cytometry assay. A) Apoptosis of RASF with transfection of anti-E2F2 siRNA or Allstars siRNA was measured, and the result is depicted in one map. B) Apoptosis of RASF with transfection of E2F2 expressing plasmids or blank plasmids (mock) was measured, and the result is depicted in one map. *p < 0.05

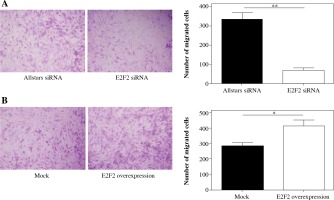
Transwell assays were performed to investigate migration ability of those 10 RASF following siRNA transfection. The assay revealed that the migration was significantly reduced in RASF with transfection of anti-E2F2 siRNA compared with the cells with transfection of Allstars siRNA (p = 0.0021). This result is shown in Figure 7A. Those 10 RASF were also transfected with E2F2-expressing plasmids. The assay showed that the migration ability was significantly increased in RASF following transfection of the E2F2-expressing plasmids compared with RASF transfected with the blank plasmids (p = 0.0456). This result is shown in Figure 7B.
Fig. 7
The effect of E2F2 expression on migration ability of RASF using transwell assays. A) The cell migration of RASF with transfection of anti-E2F2 siRNA or Allstars siRNA was measured, and the result is depicted in one map. B) The cell migration of RASF with transfection of the E2F2-expressing plasmids or the blank plasmids (mock) was measured, and the result is depicted in one map. *p < 0.05, **p < 0.01
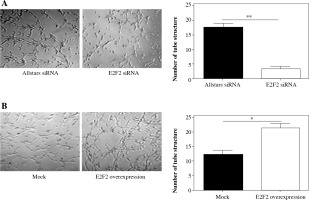
Matrigel assays were performed to observe the tube-like structure formation ability of RASF. RASF in the 3D culture generated many tube-like structures. The number of tube-like structures was significantly reduced in the anti-E2F2 siRNA-transfected RASF compared with the samples transfected with Allstars siRNA (p = 0.001). This result is shown in Figure 8A. Those 10 RASF were also transfected with E2F2-expressing plasmids. The 3D culture showed that the number of tube-like structures was significantly increased in RASF following transfection of the E2F2-expressing plasmids compared with RASF transfected with the blank plasmids (p = 0.017). This result is shown in Figure 8B.
Fig. 8
The effect of E2F2 expression on tube-like structure formation ability in RASF using Matrigel culture assay. A) Tube-like structure formation of RASF with transfection of anti-E2F2 siRNA or Allstars siRNA was measured, and the result is depicted in one map. B) Tube-like structure formation of RASF with transfection of the E2F2-expressing plasmids or the blank plasmids (mock) was measured, and the result is depicted in one map. *p < 0.05, **p < 0.01
Discussion
Previously, using transcriptome analysis, we detected increased E2F2 expression in RA synovial tissue and confirmed this finding using Western blot analysis. We also identified E2F2 expression in RA synovial cells with immunohistochemistry [9]. In the present study, we used siRNA to inhibit E2F2 expression in RASF. We found that suppressed E2F2 expression stimulated apoptosis and deduced migration and tube-like structure formation ability in RASF. On the other hand, we observed significantly increased migration and tube-like structure formation and decreased apoptosis in RASF with transfection of E2F2 overexpressing plasmids. This observation indicates that E2F2 expression mediates RASF phenotypes and activities. Abnormal synovial fibroblast proliferation, apoptosis and migration mainly contribute to RA progression. Thus, our studies suggest that the high E2F2 expression in RA synovium contributes to the pathogenic process of this disease. E2F2 promotes cell mitosis at the transcriptional level via the activation of cell cycle-related gene expression [10, 11]. High E2F2 expression has been reported in tumors, including renal cell carcinoma, thyroid cancer, non-small cell lung cancer, glioma, breast cancer, prostate cancer, colon cancer, liver cancer and glioblastoma [10-18]. Thus, some studies consider that E2F2 overexpression promotes abnormal cell proliferation and leads to tumorigenesis [15, 19, 20], which supports our observation that increased E2F2 expression lead to low apoptosis in RASF.
The present study investigated via PCR array the pathogenic mechanism of E2F2 in RASF. In this study, total RNA was extracted from 10 individual E2F2-silenced RASF samples, and the expression profile was analyzed with an inflammatory PCR array. Finally, both real-time PCR and Western blotting verified that CCR4 expression was increased in RASF with E2F2 overexpression and was decreased in RASF with inhibited E2F2 expression. This observation demonstrates that E2F2 up-regulates CCR4 expression in RASF.
CCR4 belongs to the G-protein-coupled receptor family and is a receptor for CC chemokines, such as MIP-1, RANTES, TARC and MCP-1. CCR4 has two CC chemokine ligands CCL17 (thymus- and activation-regulated chemokine) and CCL22 (macrophage-derived chemokine) [21]. Cells expressing CCR4 are found in many inflammatory diseases [21]. Many studies have demonstrated high CCR4 expression in synovial membrane and synovial fluid of RA [22]. A recent analysis with human joint fibroblast-like synoviocyte microarray profiles found that CCR4 has increased expression and might be a key gene in RA pathogenesis [23]. CCR4 expression level was correlated with activity, stage, and serological status of RA, indicating that CCR4 and its ligands could participate in spread of RA disease in the joint tissues [24]. Some reports have demonstrated that CCR4 promotes T-cell migration and proliferation [22, 25]. CCR4 was also reported to promote tumor growth and malignant behavior in breast cancer, colon cancer, gastric cancer, hepatocellular carcinoma, head and neck squamous cell carcinoma, lung cancer, melanoma and prostate cancer [26-33]. The IL-4/CCL22/CCR4/CCL17 axis was suggested to play a role to stimulate migration via RhoA/Rho-kinase signaling [28-31, 34]. The present study detected CCR4 expression in cultured RASF by real-time PCR and Western blotting and demonstrated that CCR4 expression is up-regulated by E2F2. Our study also found that inhibition of E2F2 expression retarded RASF migration and overexpression of E2F2 expression stimulated the migration. Thus, we suggest that E2F2 may activate RASF migration and might play this role by up-regulating CCR4 expression in RA pathogenesis.
In summary, the present study found that E2F2 expression regulates cell apoptosis, cell migration and tube-like structure formation in RASF, indicating that increased E2F2 expression may activate RASF to accelerate RA progression. This study also found that E2F2 stimulates CCR4 expression in RASF. These findings may be useful for understanding RA pathogenesis and signaling pathways.


