Introduction
Small cell lung cancer (SCLC), which is diagnosed in 15–20% of lung cancer patients, progresses rapidly and in many cases distant metastases are detected at the time of disease diagnosis [1–3]. The aggressive course of SCLC and its association with smoking limit the possibilities of molecularly targeted treatment, which influences the poor prognosis. The main treatment regimen in SCLC patients remains platinum-based chemotherapy in combination with etoposide, which shows satisfactory effectiveness with a high rate of 1-year survival. However, high grade toxicity of chemotherapy occurs in many patients. Lack of genetic predictive factors in SCLC limits the possibilities of personalized treatment. However, there are some promising genetic alterations that may predict efficiency and toxicity of chemotherapy [1, 2, 4].
Single nucleotide polymorphisms (SNPs) in DNA repair genes were previously reported as a predictive factors for platinum-based chemotherapy in non-small cell lung cancer (NSCLC) [2]. Moreover, polymorphic variants in DNA repair genes may interact with smoking status as risk factors of lung cancer in non-smokers but protective factors in heavy smokers who have smoked more than 15 pack years during their life [5]. Predictive value was reported for SNPs in the excision repair cross-complementation group 1 (ERCC1) gene that encodes an enzyme involved in nucleotide excision repair (NER) by removal of the damaged DNA strand. During chemotherapy, the DNA strand is damaged by cisplatin adducts. Rapid excision of platinum-DNA adducts negates the efficacy of chemotherapy and increases the ability of cell proliferation. ERCC1 polymorphisms have been extensively investigated in advanced NSCLC patients treated with platinum-based chemotherapy [2, 4, 6]. There are limited data on their importance in prognosis of SCLC patients treated with platinum-based chemotherapy [6–9].
In SCLC patients, SNPs in genes that encode topoisomerase II may play an important predictive and prognostic role. These enzymes are responsible for DNA metabolism through canalization of the ATP-dependent breakage and rejoining of the DNA double strand. Topoisomerase II occurs in α and β isoforms. Cytotoxic drugs (anthracyclines, etoposide) target topoisomerase II α (TOP2A) that regulates chromosome condensation and chromatid separation. Etoposide acts on TOP2A protein and prevents DNA replication and transcription, leading to apoptosis when DNA single- and double-strand breaks (DSBs) cannot be repaired [10]. The predictive value of TOP2A gene polymorphism was confirmed in SCLC cell lines cultured with cytostatics. However, the expression of the isoforms of topoisomerase II was considered as a prognostic factor in SCLC patients [9, 11].
Because of limited data on the clinical relevance of ERCC1 and TOP2A genetic alterations in SCLC we undertook this retrospective study to determine the influence of SNPs in TOP2A (rs34300454; rs13695; rs11540720) and ERCC1 (rs11615; rs3212986) genes on efficiency and toxicity of chemotherapy with platinum and etoposide. The SNPs that we chose showed clinical relevance in lung cancer patients.
Material and methods
Patients
The studied group included 103 Caucasian SCLC patients (65 male, 38 female, median age 65 ±7.5 years) treated in the Department of Pneumonology, Oncology and Allergology, Medical University of Lublin, between 2014 and 2017. From the whole studied group we collected demographic (age, gender, environmental/occupational exposure to carcinogens, smoking history) and clinical (stage, performance status, presence of distant metastases) data. The response to treatment was monitored according to RECIST criteria, as well as side effects such as anaemia, neutropenia or weight loss, which were monitored according to the CTCAE system [12, 13]. Detailed patient characteristics are presented in Table I. The study was approved by the Ethics Committee of the Medical University of Lublin, Poland (No. KE-0254/297/2013).
Table I
Clinical and demographic characteristics of small cell lung cancer patients
Polymorphism analysis
We obtained 10 ml of peripheral blood using the EDTA blood collection system (Sarstedt, S-MONOVETE, Germany) from each patient. The SNPs were analyzed in DNA isolated from peripheral blood leukocytes, using the QIAamp DNA Mini Kit (Qiagen, Germany) according to manufacturer procedures. For SNP genotyping we used TaqMan Genotyping Master Mix (Applied Biosystems, USA) and commercially available custom TaqMan hydrolysis probes (Applied Biosystems, USA) dedicated for research use only (RUO). The genotyping was performed in a total volume of 15 μl including 10 μl of TaqMan Genotyping Master Mix; 1 μl TaqMan hydrolysis probe (20%) and 4 μl of tested DNA (20 ng/μl). The SNPs were analyzed on an Eco Illumina (Illumina, USA) device in the following conditions – initial denaturation: 95°C – 10 min, cycling: 40 cycles: 95°C – 15 s, 60°C – 1 min. Genotyping was based on amplification of reaction primers specific for the polymorphic variants and identified by allele-specific hydrolysis probes emitting fluorescence in different bands of light (VIC/FAM). The method was validated based on analysis performed on controlled DNA with known genotype. Samples with late amplification (Ct > 35 cycles) were excluded from the analysis. Characteristics of analyzed SNPs are reported in Table II.
Table II
Characteristics of analyzed SNPs in TOP2A and ERCC1 genes
Statistical analysis
Statistical analysis was performed using Statistica version 9.0 (StatSoft, USA) and MedCalc 10 (MedCalc Software, Belgium). Differences in the frequencies of alleles between groups were tested using Fisher’s χ2 test. Odds ratios (OR) with 95% confidence intervals (95% CI) were calculated by dichotomous or trichotomous logistic regression analysis using a program that estimated correlations between alleles and genotype frequencies and clinical factors (http://ihg.gsf.de). The Mann-Whitney U-test was used for the comparison of unpaired group data. Moreover, the Kaplan-Meier method was used for calculation of survival probability. Multivariate survival analysis was analyzed using Cox’s regression model. Differences with a p value of less than 0.05 (p < 0.05) were considered significant.
Results
Genotyping of rs34300454, rs11540720 and rs13695 in the TOP2A gene indicated that the most common genotypes were C/C (98.1%; 101/103), A/A (99%; 102/103) and C/C (53.4%; 55/103), respectively. On the other hand, 64% (62/103) and 51.5% (53/103) of patients showed A/A genotype in rs11615 and A/C genotype of rs3212986 in the ERCC1 gene. The frequencies of particular genotypes in TOP2A and ERCC1 genes are presented in Table III.
Table III
Frequency of particular genotypes in TOP2A and ERCC1 genes in small cell lung cancer patients
We evaluated the association between genotypes occurrence and demographic-clinical factors in SCLC patients. We did not find a significant relationship (p > 0.05) between genotype of TOP2A and ERCC1 genes and gender, age, exposure to carcinogens (including smoking status), disease stage, presence of metastases or performance status of SCLC patients. It means that the studied genotypes of TOP2A and ERCC1 genes had no impact on demographic-clinical features, which could lead to SCLC development.
A response to chemotherapy was observed in 36.8% of SCLC patients and disease control was noted in 87.4% of examined patients (Table I). In the whole group of patients, median progression-free survival (PFS) and median overall survival (OS) were 6 months (range: 1–22 months) and 9 months (range: 1–28 months) respectively.
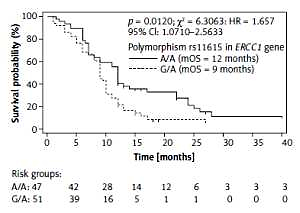
Examined SNPs did not affect the risk of early progression or the chance of response to chemotherapy in SCLC patients. Using the Kaplan-Meier method we observed that homozygous A/A genotype compared to heterozygous G/A genotype in rs11615 of the ERCC1 gene had a favourable prognostic value. Patients with G/A genotype had 9 months of OS compared to patients with A/A genotype with 12 months of OS (p = 0.01, χ² = 6.31, HR = 1.657, 95% CI: 1.0710–2.5633; Figure 1). However, rs11615 of the ERCC1 gene had no effect on PFS of SCLC patients treated with platinum-based chemotherapy (p = 0.08694, HR = 1.4647, 95% CI: 0.9483–2.2624; p model value: p = 0.0865, χ2 = 2.9379). Other examined genotypes of the TOP2A and ERCC1 genes did not significantly (p > 0.05) affect OS and PFS in SCLC patients (Table IV).
Table IV
Detailed statistical analysis
Figure 1
Overall survival of small cell lung cancer patients with different genotypes in rs11615 polymorphism of ERCC1 gene estimated by Kaplan-Meier method. A/A genotype showed favourable prognostic value (3 months longer OS) compared to G/A genotype in rs11615 of ERCC1 gene (12 vs. 9 months)
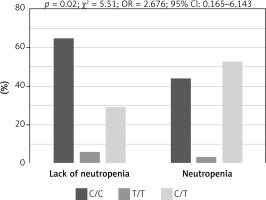
Genotypes of rs3212986 in the ERCC1 gene and rs13695 in the TOP2A gene indicated a significant impact on the risk of chemotherapy side effects (Fisher test, Table IV). Patients harbouring homozygous C/C genotype in rs3212986 of the ERCC1 gene showed significantly higher risk of anaemia during chemotherapy than heterozygous C/A patients (p = 0.045, χ² = 4.01, OR = 0.417, 95% CI: 0.175–0.991; Figure 2), whereas patients with C/C genotype in rs13695 of the TOP2A gene had significantly lower risk of neutropenia during chemotherapy than C/T heterozygous patients (p = 0.02, χ² = 5.51, OR = 2.676, 95% CI: 1.165–6.143; Figure 3).
Figure 2
Impact of genotype in rs3212986 of ERCC1 gene on risk of anaemia during chemotherapy in patients with small cell lung cancer. Patients with homozygous genotype (C/C) had significantly higher risk of anaemia during chemotherapy than heterozygous (C/A) patients
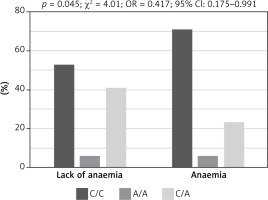
Figure 3
Impact of genotype in rs13695 of TOP2A gene on risk of neutropenia during chemotherapy in patients with small cell lung cancer. Patients with homozygous genotype (C/C) had significantly higher risk of neutropenia during chemotherapy than heterozygous (C/T) patients
Cox’s regression model indicated that homozygous genotype A/A of rs11615 in the ERCC1 gene was a single favourable prognostic factor that significantly prolonged OS in SCLC patients (p = 0.02, HR = 1.7260, 95% CI: 1.1014–2.7046; model value: p = 0.02, χ2 = 5.6773, Table IV). Moreover, the same genotype (p = 0.09, HR = 1.4647, 95% CI: 0.9483–2.2624) and younger age of SCLC onset (p = 0.08, HR = 0.6625, 95% CI: 0.4216–1.0411) insignificantly prolonged PFS (model value: p = 0.08, χ2 = 3.0151).
Discussion
In comparison to genetic mutations, SNPs are more silenced molecular alterations. However, they affect gene sequences and transcription showing changes in proteins. Sereno et al. noted that low expression of ERCC1 and TOP1 proteins was significantly associated with better response to platinum based therapy [14], whereas Chiappori et al. reported that only TOP2A expression may predict better response to chemotherapy in SCLC patients [15]. Huang et al. observed that SCLC patients with overexpression of TOP2A showed longer survival without brain metastases [16]. Hou et al. showed that high expression of TOP2A protein correlated with significantly shorter OS in NSCLC patients [11]. Moreover, the SNP profile of ERCC1 and RRM1 genes indicated clinical relevance for the response to platinum-based chemotherapy in NSCLC patients [6, 17, 18]. Based on these observations from NSCLC patients we undertook our study in order to extend the knowledge about the influence of SNPs in TOP2A and ERCC1 genes on efficiency and toxicity of chemotherapy with platinum and etoposide in SCLC, which is the first such analysis in an SCLC cohort worldwide.
We genotyped five polymorphic alterations (three in TOP2A and two in ERCC1 genes) which were previously analyzed in NSCLC patients. The SNPs that we chose may be involved in metabolism of cisplatin and etoposide also in SCLC patients [6–9]. The distribution of rs34300454 and rs11540720 in TOP2A and rs11615 in ERCC1 genes showed concordance with global studies, but the incidence of particular genotypes of rs13695 in TOP2A and rs3212986 in ERCC1 genes showed some incompatibility with literature data that might result from demographic and clinical factors [19–21]. Global data indicated that 64–74% of the healthy population worldwide harbours the C/C genotype in rs13695 [19–21], whereas this genotype was observed in 54% (56/103) of our Caucasian SCLC patients. Literature data indicate that frequencies of A/C and C/C genotypes of rs3212986 in the ERCC1 gene are 76% and 24%, respectively [19–21]. Meanwhile, 51.5% (53/103) and 48.5% (50/103) of our SCLC patients were carriers of these genotypes.
To date, the clinical impact of ERCC1 gene polymorphisms has been reported in NSCLC patients [6–8, 18, 22]. Koc et al. found that the presence of the C allele of rs11615 in the ERCC1 gene was associated with the early stage of NSCLC (p = 0.002) as well as with younger age of NSCLC patients (p = 0.04) [7]. In SCLC patients, we did not observe significant correlations between SNPs of examined genes and disease stage or patients’ age. Mlak et al. observed that distribution of alleles in rs1615 of the ERCC1 gene was insignificantly associated with duration of OS in NSCLC patients (7.5 months in carriers of C/C, 16.5 months in carriers of C/T and 13 months in carriers of T/T genotype) [18]. Zhao et al. noted that A/A genotype in rs11615 and rs3212986 of the ERCC1 gene significantly correlated with an increased risk of death in NSCLC patients [6]. Also, Gao et al. reported that presence of the A allele in rs11615 and rs3212986 of the ERCC1 gene was significantly associated with an increased risk of death in NSCLC patients [8]. Moreover, Gao et al. revealed that NSCLC patients carrying the A/A genotype in rs11615 and rs3212986 of the ERCC1 gene showed a significantly lower response rate to chemotherapy [8]. In contrast, Zhao et al. noted that NSCLC patients with A/A genotype in rs11615 and rs3212986 of the ERCC1 gene had a significantly higher response rate to chemotherapy compared to patients harbouring C/C genotype [6]. In our SCLC patients, homozygous A/A genotype in rs11615 in the ERCC1 gene showed a favourable prognostic value that was associated with significant prolongation of OS and insignificant prolongation of PFS.
Except for data on the effect of SNPs on survival, there are limited data concerning their impact on toxicity of chemotherapy in lung cancer. Kalikaki et al. found that none of the polymorphic genotypes of the ERCC1 gene (rs3212986, rs11615) correlated with haematological toxicity, including neutropenia, thrombocytopenia, anaemia, and febrile neutropenia or non-haematological toxicity in NSCLC patient [23]. In our study, SCLC patients harbouring homozygous C/C genotype in rs3212986 of the ERCC1 gene showed significantly higher risk of anaemia during chemotherapy. On the other hand, patients with C/C genotype in rs13695 of the TOP2A gene had significantly lower risk of neutropenia during chemotherapy than C/T heterozygous patients. Such observations in SCLC patients have not been published.
In conclusion, we would like to note that we, for the first time worldwide, found that some polymorphic variants of ERCC1 and TOP2A genes may have an impact on the course of SCLC, and may affect the toxicity of chemotherapy. Analysis of SNP profile may predict the early presence of serious side effects of treatment (anaemia or neutropenia) that could allow early prevention of adverse effects of chemotherapy.