Introduction
Systemic lupus erythematosus is a clinically complex disease [1]. The kidney is an organ commonly affected by systemic lupus erythematosus, and lupus nephritis is a major manifestation with high morbidity and mortality [2]. Although improvements have been gained in the pathogenesis and prognosis of lupus nephritis, the outcomes of patients remain unsatisfactory [3]. Thus, exploring novel targets for the treatment of lupus nephritis is necessary.
Circular RNAs (circRNAs) are a type of covalently closed noncoding RNAs with a lack of polyadenylation and capping in eukaryotes [4]. CircRNAs are related to the pathogenesis of multiple renal diseases, including lupus nephritis [5]. Many circRNAs are dysregulated, and play important roles in lupus nephritis [6]. For instance, circ-RNA_002453 is up-regulated in plasm of lupus nephritis patients, and can function as a biomarker for the diagnosis of lupus nephritis [7]. Moreover, circRNA human leucocyte antigen class-C (circHLA-C) is associated with the development of lupus nephritis [8]. Hsa_circ_0010957 is a circRNA derived from selenoprotein N, which is aberrantly expressed in lupus nephritis [9]. The dysregulated hsa_circ_0010957 might have an important role in lupus nephritis progression, but the exact function and mechanism are largely unknown.
MicroRNAs (miRNAs) are short noncoding RNAs (~21 nucleotides) that play key roles in the development of lupus nephritis [10]. Many miRNAs are relevant to lupus nephritis progression by way of a miRNA-mRNA network [11]. MiR-1224-5p is a miRNA associated with kidney injury [12]. However, the role of miR-1224-5p in lupus nephritis is unclear. The interleukin-1 receptor-associated kinase (IRAK) family controls the development of inflammatory diseases [13]. IRAK1 is a typical member of the IRAK family, which is implicated in lipopolysaccharide (LPS)-induced inflammatory injury in many conditions [14, 15]. Furthermore, IRAK1 contributes to the development of systemic lupus erythematosus [16, 17]. Nevertheless, the function of IRAK1 in lupus nephritis and whether IRAK1 is required for hsa_circ_0010957 and miR-1224-5p are uncertain.
In this research, we established an LPS-induced kidney injury model using HK2 cells as in previous reports [18-20]. Moreover, we measured hsa_circ_0010957, miR-1224-5p and IRAK1 expression in LPS-challenged cells, and explored the function of hsa_circ_0010957 in LPS-induced cell damage. Additionally, we evaluated the relationship of hsa_circ_0010957, miR-1224-5p and IRAK1.
Material and methods
Cell culture and exposure to lipopolysaccharide
Human renal proximal tubular cell line HK2 cells were purchased from Procell (Wuhan, China), and were cultured in minimum Eagle’s medium (Procell) with 10% fetal bovine serum (HyClone, Logan, UT, USA) and 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C and 5% CO2.
To mimic lupus nephritis-like injury, HK2 cells were exposed to various doses (1, 5 and 10 µg/ml) of LPS (purity ≥ 99%; Solarbio, Beijing, China) for 24 h. Cells without LPS treatment were used as a control.
Quantitative reverse transcription polymerase chain reaction
Total RNA was extracted using Trizol (Beyotime, Shanghai, China). To detect RNA expression level, 1 µg RNA was reversely transcribed into cDNA with a M-MLV Reverse Transcriptase kit (Thermo Fisher Scientific) or TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher Scientific). The cDNA was mixed with SYBR (Vazyme, Nanjing, China) and specific primers (Genscript, Nanjing, China) for quantitative reverse transcription polymerase chain reaction (qRT-PCR). The primer sequences were as follows: hsa_circ_0010957 (sense, 5'-GCTGCCCTGTCACTCCTCTA-3’; antisense, 5’-CACGGAGGTGATGTCCAAG-3’), IRAK1 (sense, 5’-CAGTTCGCCGCCCTGAT-3’; antisense, 5’-CTGGAAAAGCTGGGGAGAGG-3’), miR-1224-5p (sense, 5’-GCCGAGGTGAGGACTCGGGA-3’; antisense, 5’-CAGTGCGTGTCGTGGAGT-3’), U6 (sense, 5’-CTCGCTTCGGCAGCACA-3’; antisense, 5’-AACGCTTCACGAATTTGCGT-3’), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (sense, 5’-CCTGTTCGACAGTCAGCCG-3’; antisense, 5’-GAGAACAGTGAGCGCCTAGT-3’). U6 or GAPDH served as a housekeeping gene, and relative RNA level was calculated via the 2-△△Ct method [21].
Cell transfection
The Hsa_circ_0010957 overexpression vector was synthesized using the pCD5-ciR vector (Geneseed, Guangzhou, China), with the vector alone as a negative control (circ-NC). The IRAK1 overexpression vector was based on the pcDNA3.1 vector (Thermo Fisher Scientific), and the empty vector served as a negative control (vector). The siRNA of hsa_circ_0010957 (si-hsa_circ_0010957, 5’-AUUGAUGUGAUGGACGUUAGU-3’), negative control of siRNA (si-NC) (5’-AAGACAUUGUGUGUCCGCCTT-3’), miR-1224-5p mimic (5’-GUGAGGACUCGGGAGGUGG-3’), mimic negative control (NC, 5’-UUCUCCGAACGUGUCACGU-3’), miR-1224-5p inhibitor (anti-miR-1224-5p, 5’-CCACCUCCCGAGUCCUCAC-3’), and inhibitor negative control (anti-NC, 5’-UGAGCUGCAUAGAGUAGUGAUUA-3’) were obtained from GenePharma (Shanghai, China). Cell transfection was conducted in HK2 cells using Lipofectamine 2000 (Thermo Fisher Scientific) for 24 h. The transfection efficacy was identified via qRT-PCR or western blot. Non-transfected cells were regarded as a blank group.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
1 × 104 HK2 cells were added to 96-well plates overnight, and then exposed to LPS for 24 h. Then, cell medium was replaced with fresh medium, adding 0.5 mg/ml MTT (Solarbio). Following incubation for 4 h, the medium was discarded, and 150 µl of dimethyl sulfoxide (Solarbio) was injected. The absorbance was detected at 570 nm by a microplate reader (Potenov, Beijing, China). Relative cell viability was normalized to the control group (100%).
Flow cytometry
Cell apoptosis was examined via flow cytometry with an Annexin V-FITC apoptosis kit (Yeasen, Shanghai, China). 2 × 105 HK2 cells were placed in 12-well plates, and stimulated with LPS for 24 h. Next, cells were harvested and interacted with Annexin V binding buffer, followed by dyeing with Annexin V-FITC and propidium iodide. The stained cells were detected with a flow cytometer (Agilent, Beijing, China). The apoptotic rate was shown as a percentage of cells in the right lower and upper quadrants.
Enzyme-linked immunosorbent assay
The release of pro-inflammatory cytokines was measured via enzyme-linked immunosorbent assay (ELISA). 2 × 105 HK2 cells were added to 12-well plates, and exposed to LPS for 24 h. Next, medium was harvested, and 50 µl samples were used for detection of tumor necrosis factor α (TNF-α), interleukin (IL)-8, IL-1β and IL-6 levels using the specific TNF-α, IL-8, IL-1β or IL-6 ELISA kit (R&D Systems, Minneapolis, MN, USA). The limit of detection (LOD) for TNF-α, IL-8, IL-1β or IL-6 ELISA kit was 6.23, 7.5, 1.0 or 0.7 pg/ml, respectively. The absorbance was examined at 450 nm using a microplate reader.
Measurement of reactive oxygen species, lactate dehydrogenase and superoxide dismutase levels
For reactive oxygen species (ROS) level analysis, 1 × 104 HK2 cells were placed into 96-well plates and exposed to LPS for 24 h. Next, cells were incubated with a cellular ROS assay kit (Abcam, Cambridge, MA, USA) according to the instructions of the manufacturer. The fluorescence signal was measured via a fluorescent microplate reader (Molecular Devices, Sunnyvale, CA, USA) with excitation/emission at 485/535 nm. The relative ROS activity was normalized to the control group.
For lactate dehydrogenase (LDH) level analysis, 1 × 104 HK2 cells were seeded into 96-well plates and treated with LPS for 24 h. Medium was harvested, and LDH release was detected using an LDH assay kit (Abcam) following the manufacturer’s instructions. The absorbance was detected at 450 nm with a microplate reader. Relative LDH level was normalized to the control group.
For superoxide dismutase (SOD) level assay, 2 × 105 HK2 cells were added to 12-well plates, and stimulated with LPS for 24 h. Next, cells were lysed and used for detection of SOD level using an SOD activity assay kit (Abcam) according to the manufacturer’s instructions. The absorbance was examined at 450 nm through a microplate reader. Relative SOD level was normalized to the control group.
Western blot
HK2 cells were lysed in RIPA buffer (Beyotime), and the cell lysates were centrifugated at 14,000 g for 1 min. The supernatants were obtained, and protein concentration was detected with a BCA kit (Amyjet, Wuhan, China). A 20 µg protein sample was separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membrane (Solarbio). The membrane was blocked in 5% bovine serum albumin (Solarbio). Subsequently, the membrane was incubated with antibody against B-cell lymphoma-2 (Bcl-2) (ab59348, 1 : 500 dilution, Abcam), Bcl-2-associated X (Bax) (ab263897, 1 : 2000 dilution, Abcam), cleaved caspase 3 (C-caspase 3) (ab2302, 1 : 1000 dilution, Abcam), IRAK1 (ab238, 1 : 2000 dilution, Abcam), or GAPDH (ab245355, 1 : 5000 dilution, Abcam) overnight and IgG labeled by horseradish peroxidase (HRP) (ab6721, 1 : 8000 dilution, Abcam) for 2 h. Next, the blots were interacted with ECL reagent (Yeasen), and then examined by Image J v1.8 software (NIH, Bethesda, MD, USA). GAPDH functioned as a loading reference, and relative protein expression was normalized to the control group.
Dual-luciferase reporter analysis and RNA immunoprecipitation
The target association between hsa_circ_0010957 and miR-1224-5p or miR-1224-5p and IRAK1 was predicted via starBase. The wild-type luciferase reporter vectors hsa_circ_0010957-wt and IRAK1-wt were constructed via cloning the sequence of hsa_circ_0010957 or IRAK1 3’UTR containing the miR-1224-5p complementary sequence into the pmirGLO vector (Promega, Madison, WI, USA). The mutant-type luciferase reporter vectors hsa_circ_0010957-mut and IRAK1-mut were formed using the sequence with mutant seed sites (AGUCCUCA to UCAGAAGU in hsa_circ_0010957-mut; CGAGUCCUCA to GCUCAGUAGU in IRAK1-mut). These luciferase reporter vectors were transfected into HK2 cells together with the miR-1224-5p mimic or NC for 24 h. Next, the luciferase activity was detected via a luciferase analysis kit (Promega).
The Magna RIP kit (Sigma, St. Louis, MO, USA) was used for RNA immunoprecipitation (RIP) analysis. 1 × 107 HK2 cells were lysed and incubated with magnetic beads precoated with antibody against Ago2 (anti-Ago2) or IgG (anti-IgG) for 8 h at 4°C. Next, the beads were incubated with Trizol, and levels of hsa_circ_0010957, miR-1224-5p and IRAK1 enriched on the beads were detected via qRT-PCR. Anti-IgG was used as a negative control, and Input was used as a positive control.
Statistical analysis
GraphPad Prism 6 (GraphPad Inc., La Jolla, CA, USA) was processed for statistical analysis. The experiments were repeated 3 times, and each reaction contained 3 replicates. Results are shown as mean ± standard deviation (SD). The difference was evaluated via ANOVA followed by the Tukey post hoc test. It was significant at p < 0.05.
Results
Hsa_circ_0010957 expression is up-regulated and miR-1224-5p level is down-regulated in LPS-challenged HK2 cells
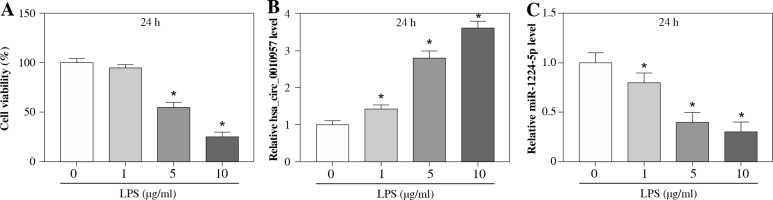
To analyze whether hsa_circ_0010957 and miR-1224-5p were associated with LPS-induced damage in HK2 cells, HK2 cells were stimulated by various doses (0, 1, 5 and 10 µg/ml) of LPS for 24 h. As displayed in Figure 1A, HK2 cell viability was reduced via treatment with LPS in a dose-dependent pattern, and the change was significant when treated with 5 and 10 µg/ml of LPS. Moreover, hsa_circ_0010957 and miR-1224-5p abundances were detected in LPS-challenged HK2 cells. The results showed that hsa_circ_0010957 abundance was progressively enhanced via exposure to LPS in a dose-dependent pattern, while miR-1224-5p level was reduced (Fig. 1B, C). These results indicated that the dysregulated hsa_circ_0010957 and miR-1224-5p might be involved in LPS-induced HK2 cell injury. 5 µg/ml of LPS that induced obvious viability reduction was chosen as an exposure dose in further study.
Fig. 1
Hsa_circ_0010957 and miR-1224-5p expression in LPS-challenged HK2 cells. A) Cell viability was measured by MTT in HK2 cells after treatment with 0, 1, 5 or 10 μg/ml of LPS for 24 h; n = 3. B, C) Hsa_circ_0010957 and miR-1224-5p levels were measured via qRT-PCR in HK2 cells after treatment with 0, 1, 5 or 10 μg/ml of LPS for 24 h; n = 3. *p < 0.05
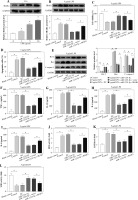
Hsa_circ_0010957 knockdown attenuates LPS-induced apoptosis, inflammatory response and oxidative injury in HK2 cells
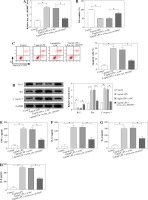
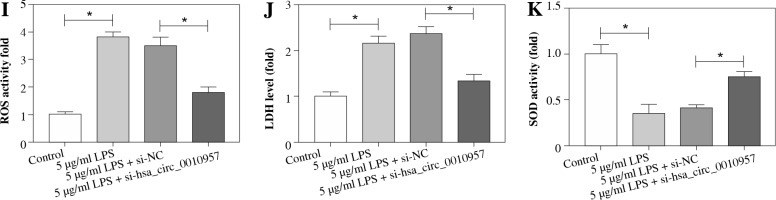
To study the effect of hsa_circ_0010957 on LPS-induced HK2 cell injury, cells were transfected with si-NC or si-hsa_circ_0010957, and treated with 5 µg/ml of LPS for 24 h. As shown in Figure 2A, hsa_circ_0010957 abundance in HK2 cells was enhanced 3-fold by exposure to 5 µg/ml of LPS, and it was effectively decreased via transfection of si-hsa_circ_0010957. MTT analysis revealed that HK2 cell viability was decreased 50% by stimulation of LPS, which was restored via hsa_circ_0010957 down-regulation (Fig. 2B). Moreover, a higher apoptotic rate of HK2 cells was induced via LPS treatment, which was weakened by hsa_circ_0010957 knockdown (Fig. 2C). In addition, the anti-apoptotic protein Bcl-2 and pro-apoptotic protein Bax and C-caspase 3 levels were detected in HK2 cells. LPS markedly reduced Bcl-2 abundance and increased levels of Bax and C-caspase 3, and this effect was mitigated by hsa_circ_0010957 silence (Fig. 2D). Furthermore, LPS significantly promoted the release of TNF-α, IL-1β, IL-6 and IL-8 in HK2 cells, which was weakened via hsa_circ_0010957 knockdown (Fig. 2E-H). Also, LPS evidently enhanced ROS and LDH levels but decreased the SOD level, which was reversed via hsa_circ_0010957 interference (Fig. 2I-K). These data indicated that hsa_circ_0010957 silence mitigated LPS-induced HK2 cell damage.
Fig. 2
The influence of hsa_circ_0010957 on LPS-induced damage in HK2 cells. A) Hsa_circ_0010957 expression was examined via qRT-PCR in HK2 cells with transfection of si-NC or si-hsa_circ_0010957 after treatment with 5 μg/ml of LPS for 24 h; n = 3. B) Cell viability was measured via MTT in HK2 cells with transfection of si-NC or si-hsa_circ_0010957 after exposure to 5 μg/ml of LPS for 24 h; n = 3. C) Apoptotic rate was measured by flow cytometry in HK2 cells with transfection of si-NC or si-hsa_ circ_0010957 following stimulation with 5 μg/ml of LPS for 24 h; n = 3. D) Apoptotic-related protein abundances were detected via western blot in HK2 cells with transfection of si-NC or si-hsa_circ_0010957 following stimulation with 5 μg/ml of LPS for 24 h; n = 3. E-H) The inflammatory cytokine levels were measured by ELISA in HK2 cells with transfection of si-NC or si-hsa_circ_0010957 following exposure to 5 μg/ml of LPS for 24 h; n = 3. *p < 0.05 I-K) ROS, LDH and SOD levels were examined in HK2 cells with transfection of si-NC or si-hsa_circ_0010957 after exposure to 5 μg/ml of LPS for 24 h; n = 3. <p < 0.05
MiR-1224-5p is targeted and regulated by hsa_circ_0010957 in HK2 cells
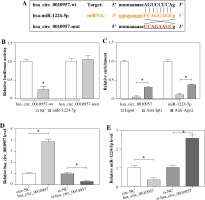
To explore the association of hsa_circ_0010957 and miR-1224-5p, we searched whether there was a binding site between them using starBase. The predicted binding sequence is shown in Figure 3A. Furthermore, dual-luciferase reporter and RIP analyses were conducted to identify the target interaction of hsa_circ_0010957 and miR-1224-5p. MiR-1224-5p overexpression reduced luciferase activity by 76% in the hsa_circ_0010957-wt group, but it displayed little influence on the luciferase activity in the hsa_circ_0010957-mut group (Fig. 3B). Moreover, RIP analysis revealed that the amount of hsa_circ_0010957 and miR-1224-5p was enriched on the same complex via Ago2 RIP (Fig. 3C). Additionally, the influence of hsa_circ_0010957 on miR-1224-5p abundance was assessed in HK2 cells. The overexpression or knockdown efficacy of hsa_circ_0010957 is shown in Figure 3D. MiR-1224-5p level was reduced 67% by hsa_circ_0010957 overexpression, and increased 2.58-fold via hsa_circ_0010957 knockdown (Fig. 3E). These results suggested that hsa_circ_0010957 could target miR-1224-5p in HK2 cells.
Fig. 3
Target association of hsa_circ_0010957 and miR-1224-5p. A) Binding sequence of hsa_circ_0010957 and miR-1224-5p. B) Luciferase activity was detected in HK2 cells with transfection of hsa_circ_0010957-wt or hsa_ circ_0010957-mut and miR-1224-5p mimic or NC; n = 3. C) Hsa_circ_0010957 and miR-1224-5p levels were detected after RIP; n = 3. D, E) Hsa_circ_0010957 and miR-1224-5p abundances were examined in HK2 cells with transfection of circ-NC, hsa_circ_0010957 overexpression vector, si-NC or si-hsa_circ_0010957; n = 3. *p < 0.05
MiR-1224-5p knockdown mitigates the effect of hsa_circ_0010957 silence on LPS-induced apoptosis, inflammatory response and oxidative injury in HK2 cells
To study whether miR-1224-5p was associated with hsa_circ_0010957-mediated regulation of LPS-induced HK2 cell damage, cells were transfected with si-NC, si-hsa_circ_0010957, si-hsa_circ_0010957 + anti-NC or anti-miR-1224-5p, and then stimulated with 5 µg/ml of LPS for 24 h. In the presence of LPS, miR-1224-5p abundance was elevated by hsa_circ_0010957 knockdown, which was reduced via transfection of anti-miR-1224-5p (Fig. 4A). Furthermore, miR-1224-5p knockdown abolished the protective effect of hsa_circ_0010957 silence on cell viability in LPS-challenged HK2 cells (Fig. 4B). In addition, miR-1224-5p down-regulation attenuated knockdown of the hsa_circ_0010957-mediated suppressive role in cell apoptosis (Fig. 4C-E). Moreover, miR-1224-5p knockdown delayed the inhibitory role of hsa_circ_0010957 interference in secretion of TNF-α, IL-1β, IL-6 and IL-8 in HK2 cells treated with LPS (Fig. 4F-I). Additionally, silencing of miR-1224-5p abrogated knockdown of hsa_circ_0010957-mediated reduction of ROS and LDH levels and promotion of SOD level (Fig. 4J-L). These results indicated that miR-1224-5p was required for hsa_circ_0010957 in regulating LPS-induced HK2 cell injury.
Fig. 4
The influence of miR-1224-5p on hsa_circ_0010957-mediated regulation of LPS-induced injury in HK2 cells. MiR-1224-5p expression (A), cell viability (B), apoptotic rate (C), apoptotic-related protein levels (D, E), inflammatory cytokine levels (F-I), ROS, LDH and SOD levels (J-L) were measured in HK2 cells with transfection of si-NC, si-hsa_circ_0010957, si-hsa_circ_0010957 + anti-NC or anti-miR-1224-5p following stimulation with 5 μg/ml of LPS for 24 h; n = 3. *p < 0.05
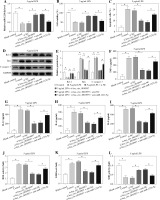
IRAK1 is targeted and regulated via the hsa_circ_0010957/miR-1224-5p axis in HK2 cells
To further explore the potential target of the hsa_circ_0010957/miR-1224-5p axis, the target of miR-1224-5p was predicted using starBase software. IRAK1 was a candidate, and the complementary sequence between miR-1224-5p and IRAK1 is shown in Figure 5A. To identify the binding correlation between them, we constructed the luciferase reporter vectors IRAK1-wt and IRAK1-mut. As displayed in Figure 5B, the luciferase activity of IRAK1-wt was reduced 77% by miR-1224-5p overexpression, but the luciferase activity of IRAK1-mut was not changed. Additionally, miR-1224-5p and IRAK1 could be enriched on the same complex by Ago2 RIP (Fig. 5C). The transfection efficacy of miR-1224-5p mimic or anti-miR-1224-5p is presented in Figure 5D. Furthermore, IRAK1 protein level was negatively regulated by miR-1224-5p (Fig. 5E). Also, the influence of hsa_circ_0010957/miR-1224-5p axis on IRAK1 expression was detected. The results showed that IRAK1 protein expression was evidently decreased by hsa_circ_0010957 silence, which was restored via miR-1224-5p knockdown (Fig. 5F). These results indicated that hsa_circ_0010957 could regulate IRAK1 via miR-1224-5p.
Fig. 5
Target relationship of miR-1224-5p and IRAK1. A) Binding site of miR-1224-5p and IRAK1. B) Luciferase activity was examined in HK2 cells with transfection of IRAK1-wt or IRAK1-mut and miR-1224-5p mimic or NC; n = 3. C) MiR-1224-5p and IRAK1 enrichment levels were examined after RIP; n = 3. D, E) MiR-1224-5p and IRAK1 levels were examined in HK2 cells with transfection of NC, miR-1224-5p mimic, anti-NC or anti-miR-1224-5p; n = 3. F) IRAK1 protein abundance was detected in HK2 cells with transfection of si-NC, si-hsa_circ_0010957, si-hsa_ circ_0010957 + anti-NC or anti-miR-1224-5p; n = 3. *p < 0.05
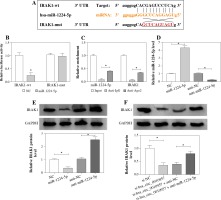
MiR-1224-5p mitigates LPS-induced apoptosis, inflammatory response and oxidative injury via regulating IRAK1 in HK2 cells
IRAK1 protein expression was markedly increased in HK2 cells after treatment with LPS for 24 h (Fig. 6A). To study the function of miR-1224-5p and explore whether it required IRAK1, HK2 cells were transfected with NC, miR-1224-5p mimic, miR-1224-5p mimic + vector or IRAK1 overexpression vector, and treated with 5 µg/ml of LPS for 24 h. In LPS-treated cells, IRAK1 expression was evidently decreased by miR-1224-5p addition, which was up-regulated via introduction of the IRAK1 overexpression vector (Fig. 6B). MTT analysis showed that miR-1224-5p overexpression attenuated LPS-induced viability inhibition, and this effect was abolished via IRAK1 restoration (Fig. 6C). Moreover, miR-1224-5p overexpression weakened LPS-induced apoptosis by decreasing the apoptotic rate and regulating apoptotic-related protein levels, which was reversed via IRAK1 addition (Fig. 6D, E). In addition, miR-1224-5p overexpression alleviated LPS-induced secretion of TNF-α, IL-1β, IL-6 and IL-8, which was overturned via IRAK1 up-regulation (Fig. 6F-I). Moreover, miR-1224-5p overexpression attenuated LPS-induced promotion of ROS and LDH levels and reduction of SOD level, which was abrogated via IRAK1 overexpression (Fig. 6J-L). These data suggested that miR-1224-5p could mitigate LPS-induced HK2 cell injury by regulating IRAK1.
Fig. 6
The influence of miR-1224-5p and IRAK1 on LPS-induced injury in HK2 cells. A) IRAK1 expression was detected in HK2 cells after treatment with 0, 1, 5 or 10 μg/ml of LPS for 24 h. IRAK1 abundance (B), cell viability (C), apoptotic rate (D), apoptotic-related protein levels (E), inflammatory cytokine levels (F-I), ROS, LDH and SOD levels (J-L) were examined in HK2 cells with transfection of NC, miR-1224-5p mimic, miR-1224-5p mimic + vector or IRAK1 overexpression vector following exposure to 5 μg/ml of LPS for 24 h; n = 3. *p < 0.05
Discussion
Lupus nephritis is a form of glomerulonephritis accompanied by systemic lupus erythematosus [22]. Many circRNAs are abnormally expressed, and may contribute to the diagnosis and treatment of lupus nephritis [5]. This study aimed to find a potential circRNA as a target for the treatment of lupus nephritis. Here we established a kidney injury model using LPS-challenged HK2 cells. Moreover, we found that hsa_circ_0010957 knockdown could attenuate LPS-induced injury, which was associated with the miR-1224-5p/IRAK1 axis.
Previous research showed that hsa_circ_0010957 expression was enhanced in lupus nephritis [9], implying that this circRNA may play an important role in lupus nephritis development. Here we found that hsa_circ_0010957 level was up-regulated in LPS-challenged HK2 cells, indicating that increased hsa_circ_0010957 might be involved in LPS-induced injury. Bcl-2 protein protects cells from programmed cell death (apoptosis), while Bax as a homologous binding partner of BCL-2 can promote cell apoptosis [23]. The activation of caspase 3 (C-caspase 3) mediates apoptosis and is responsible for killing cells [24]. Our study showed that hsa_circ_0010957 knockdown mitigated LPS-induced HK2 cell apoptosis by increasing Bcl-2 and decreasing Bax and C-caspase 3. Moreover, the inflammatory cytokines have important roles in lupus nephritis progression [25]. TNF-α, IL-1β, IL-6 and IL-8 are four key cytokines in lupus nephritis development [26, 27]. By detecting release of these cytokines using ELISA, we found that hsa_circ_0010957 silence weakened LPS-induced inflammatory injury in HK2 cells. In addition, oxidative injury is another factor for lupus nephritis [28, 29]. Through detecting the levels of oxidative stress-associated ROS and LDH and anti-oxidative SOD activity, we found that hsa_circ_0010957 silence attenuated LPS-induced oxidative injury. Collectively, hsa_circ_0010957 knockdown alleviated LPS-induced HK2 cell injury, indicating that hsa_circ_0010957 might be used as a target for treatment of lupus nephritis.
MiR-1224-5p expression was increased in acute kidney injury induced by ischemia-reperfusion, suggesting that miR-1224-5p might be associated with kidney injury [12]. However, lowly expressed miR-1224-5p was measured in LPS-challenged HK2 cells. We hypothesized that it might be due to the different microenvironments induced via ischemia-reperfusion or LPS. Niu et al. suggested that miR-1224-5p has an anti-inflammatory role in LPS-mediated inflammatory responses [30]. Here we also found that miR-1224-5p inhibited LPS-mediated inflammatory cytokine release in HK2 cells. Furthermore, our results showed that miR-1224-5p attenuated LPS-induced apoptosis and oxidative injury, although miR-1224-5p may have a promoting role in oxidative stress in acute liver injury [31, 32]. We hypothesized that the function of miR-1224-5p in response to oxidative stress might have tissue specificity. Thus, we thought miR-1224-5p could exhibit a protective function in LPS-induced HK2 cell injury. More importantly, we found that miR-1224-5p was targeted and negatively regulated via hsa_circ_0010957. Moreover, miR-1224-5p knockdown abolished the influence of hsa_circ_0010957 silence on LPS-induced HK2 cell damage. In this way, hsa_circ_0010957 could regulate LPS-induced HK2 cell injury via modulating miR-1224-5p.
Next, we explored and confirmed that IRAK1 was targeted by miR-1224-5p. Increasing evidence has indicated that IRAK1 promotes LPS-induced inflammatory injury in multiple cell lines, such as nucleus pulposus and pulmonary cells [14, 15]. Furthermore, IRAK1 contributed to the inflammatory injury in systemic lupus erythematosus [16, 17]. Additionally, IRAK1 was associated with kidney injury in systemic lupus erythematosus [33]. In this research, we found that IRAK1 facilitated LPS-induced injury in HK2 cells via reversing the effect of miR-1224-5p. Also, we found that hsa_circ_0010957 could regulate IRAK1 expression, which was weakened by miR-1224-5p, indicating that hsa_circ_0010957 could target IRAK1 by competitively binding with miR-1224-5p. In this way, hsa_circ_0010957 regulated LPS-induced HK2 cell injury via the miR-1224-5p/IRAK1 axis.
Conclusions
In conclusion, hsa_circ_0010957 silence mitigated LPS-induced apoptosis, the inflammatory response and oxidative injury in HK2 cells, possibly via the miR-1224-5p/IRAK1 axis. This study indicates the role of hsa_circ_0010957 in regulating LPS-induced HK2 cell injury, and suggests that hsa_circ_0010957 may function as a target for the treatment of lupus nephritis.


