Introduction
Allergens derived from American cockroach (Periplaneta Americana – Per a) have been recognized as major triggering factors for allergic respiratory symptoms [1]. Epidemiological studies revealed that the prevalence of cockroach (CR) allergy ranges from 17% to 41 % in the USA [2, 3]. In China, in total 25.7% of allergy patients are skin prick test (SPT) positive to the American CR [4]. Moreover, CR allergens are found in 11% to 98% of dust samples collected from 9 cities across the southern and tropical regions of China [5].
CR allergens consist of a wide group of proteins with diverse structures and biological functions. At least twenty-two immunoglobulin E (IgE) binding allergens were identified in American CR [6], but only a few of these allergens including Per a 1 (enzyme related with digestion) [7], Per a 2 (aspartic protease) [8], Per a 3 (arylphorin) [9], Per a 4 (lipocalin) [10], Per a 5 [11], Per a 6 (troponin-C) [12], Per a 7 (tropomyosin) [13], Per a 9 (arginine kinase) [14, 15], Per a 10 (serine protease) [16, 17], Per a 11 (α-amylase) [18] and Per a 12 (chitinase) [18] are characterized. Among these, Per a 9 is an arginine kinase, purified from the American CR extract by the monoclonal antibody based-affinity chromatography reacting with IgE in sera of all CR allergic Thai patients [14]. Moreover, Per a 9 was successfully expressed in E. coli and purified by 6-His-tag purification system [15, 19]. The recombinant allergen was identified as its affinity to IgE antibodies from the CR-allergic patient sera by western blotting and ELISA [15, 19].
Many, but not all allergens expressed from cDNA have shown a considerable IgE binding reactivity that seems to be comparable to their natural counterparts. The majority of these recombinant allergens are produced in E. coli, but unfortunately, the amount and/or reactivity is sometimes reduced when the allergen is purified and subjected to immunological and biochemical assays [20]. To overcome some of these problems, eukaryotic expression systems such as yeast and baculovirus in insect cells have been used [21]. Nowadays, several insect cell lines and a variety of baculovirus-based expression systems are available for the production of pharmaceutically relevant proteins. Currently, there is no report about CR allergen Per a 9 expressed in insect cells using a baculovirus expression system. The aim of the present study is to generate recombinant Per a 9 (rPer a 9) using a eukaryotic expression system (baculovirus-infected insect cells) and characterize its biochemical and immunologic properties.
Material and methods
Patients and samples
A total of 16 allergic rhinitis patients with positive STP (allergens were supplied by ALK-Abelló, Inc., Denmark) and positive serum IgE test to American CR extract (by using Immun<sup>o</sup>CAP assay [Pharmacia Diagnostics AB, Uppsala, Sweden]), and 6 healthy controls (HCs) were recruited in the study. The informed consent from each volunteer according to the Declaration of Helsinki and agreement with the ethical committee of the First Affiliated Hospital of Nanjing Medical University was obtained. Serum (4 ml) from peripheral venous blood was collected from each patient and HC for Western blot analysis.
Expression and purification of Per a 9 in baculovirus-infected insect cells
The cDNAs encoding Per a 9 was prepared as previously reported [15]. The identified Per a 9 gene was then subcloned into pFastBac1 vector (Novagen, Madison, Wisc., USA) using EcoR I and Sal I sites and the resulted construct was transformed into E. coli strain DH10Bac to generate recombinant bacmid. The positive colonies were selected and followed by PCR identification. The recombinant Bacmid was transfected into sf-9 cells by using Cellfectin (Invitrogen Corporation, Carlsbad, USA), and incubated in HyQ liquid medium (HyClone, Logan, USA) for 5-7 days at 27°C until the cells got swollen. The supernatant was collected and designated as P1 viruses. Sf-9 cells were then infected with P1 viruses at the MOI of 1, and collected when they got swollen. The cell pellet was lysed, and centrifuged at 12,000 × γ at 4°C for 20 min before the supernatant being collected. The supernatant was loaded on the Nickel column (Genscript, Nanjing, China), washed with running buffer containing 50 mM Tris-HCl, 300 mM NaCl and 5% glycerol (pH 8.0), and eluted with elution buffer containing 50 mM Tris-HCl, 300 mM NaCl, 250 mM imidazole and 5% glycerol (pH 8.0). The eluted fractions were obtained and identified as Per a 9.
Immunoreactivity of human sera with Per a 9
A 96-well plate was coated with 100 μl/well of Per a 9 (10 μg/ml) in coating buffer (50 mM sodium carbonate buffer, pH 9.6) and incubated at 4°C overnight. Human serum samples (1 : 20 dilution in PBS-Tween with 2% BSA) were then added to the plates for 2 h at room temperature. After IgE binding, plates were incubated with horseradish peroxidase-labeled goat anti-human IgE (1 : 2500 dilution) (KPL, Inc., Maryland, USA) and the color was developed with tetramethylbenzidine peroxidase substrate. The plates were read on a microplate reader at absorbance of 405 nm. The cutoff value was calculated as the mean of the negative controls plus 2 SDs.
Immunoblot analysis of IgE reactivity
Immunoblot assay for the IgE binding activity of Per a 9 to the sera of patients with American CR allergy was performed as previously described [22, 23]. Recombinant Per a 9 (5 μg) was added to a SDS-PAGE (gel concentration of 12%) under reducing conditions and then transferred to nitrocellulose membranes. The nitrocellulose membranes were incubated with the sera of the patients with American CR allergy (1 : 5 to 1 : 20 in PBS-Tween) for 90 min. Following rinsing with PBS, the membranes were incubated with peroxidase-labeled anti-human IgE monoclonal antibody. The positive protein bands were visualized by incubating the membranes with tetramethyl-benzidine peroxidase substrate. Two sera collected from HCs were used as negative controls in the experiment.
Basophil activation test
Expression of CD63 and CCR3 on basophil surface has been considered as the indicator of basophil activation [24, 25]. Briefly, peripheral blood mononucleated cells (PBMC) from 4 healthy volunteers were separated by Ficoll-Paque density gradient, and treated with 10 ml LS (a solution containing 1.3 M NaCl, 0.005 M KCl and 0.01 lactic acid, pH 3.9) for 2 min at 8°C. After neutralization with 12% Tris (pH 10.9), non-specific IgE on basophils was stripped off. Then the cells were passively sensitized with sera of the patients with American CR allergy or HCs (n = 4, 1 in 10 dilution, 2 h at 37°C) as described previously [26]. The sensitized cells were then challenged with various concentrations of Per a 9 for 15 min at 37°C. CR3-PE-labelled antibody (eBioscience Inc. San Diego, CA, USA) and anti-human CD63-FITC antibody (Invitrogen Corporation, Camarillo, CA, USA) were added to cells. Flow cytometry analysis of CD63 and CCR3 was performed at 488 nm on a FACSAria flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) and analyzed by FACSDiva software.
Results
Expression and purification of Per a 9 in baculovirus-infected insect cells
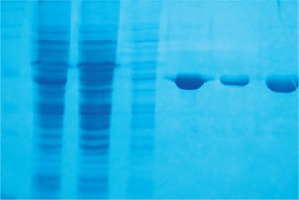
The Per a 9 encoding gene was subcloned into pFastBac1 vector and transformed into E. coli strain DH10Bac to generate recombinant bacmid. The recombinant bacmid was then transfected into Sf-9 cells to generate the baculovirus. The Per a 9 protein was expressed in Sf-9 cells and purified by Ni column. More than 6 mg recombinant Per a 9 was obtained from 2 l cell culture. The purity of the purified Per a 9 was identified by SDS-PAGE. It showed a single band with an apparent molecular weight of 42 kDa (Fig. 1).
Fig. 1
SDS-PAGE analysis of purification of Per a 9 in baculovirus-infected insect cells through Ni column. Lane M: protein standard. Lane 1: the cytosol. Lane 2: flow through. Lane 3: wash with a buffer containing 50 mM Tris-HCl, 300 mM NaCl, 5% glycerol, pH 8.0. Lane 4: elute with a buffer containing 50 mM Tris-HCl, 300 mM NaCl, 5% glycerol, 20 mM imidazole, pH 8.0. Lane 5: elute with a buffer containing 50 mM Tris-HCl, 300 mM NaCl, 5% glycerol, 50 mM imidazole, pH 8.0. Lane 6: elute with a buffer containing 50 mM Tris-HCl, 300 mM NaCl, 5% glycerol, 250 mM imidazole, pH 8.0. The arrow represents Per a 9 protein
Immuno-reactivity to IgE
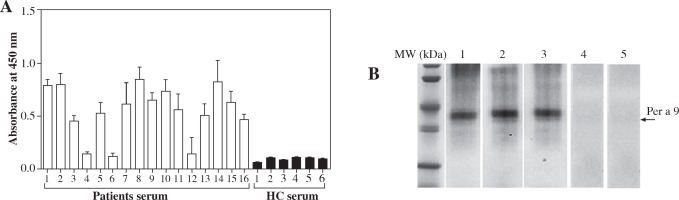
In order to determine the allergenicity of Per a 9, we examined the ability of Per a 9 to bind IgE in the sera of the patients with American CR allergy by a direct ELISA technique. The results showed that 13 out of 16 (81.3%) sera from these patients reacted to Per a 9 (Fig. 2A). The IgE reactivity of Per a 9 in the sera from the Per a 9 positive patients was increased 8.3-fold in comparison with the sera from HCs. IgE binding activity of Per a 9 in a representative group of 3 patients and 2 HCs were assessed by Western blot and were illustrated in Figure 2B. IgE binding bands appeared clear with Per a 9 reacting to serum from the patients with American CR allergy. But, Per a 9 did not react to the sera from the HCs.
Fig. 2
A) Analysis of specific IgE reactivity of recombinant Per a 9 by direct ELISA. The sera were collected from the patients with American CR allergy and HCs. The values shown are mean ±SE for the triplicate experiments. B) Western blot analysis of IgE reactivity to Per a 9 sera from the patients with American CR allergy. Lanes 1-3: Per a 9 reacted with the serum from patients 8, 9, 10. Lanes 4-5: Per a 9 reacted with the serum from control 1 and 2
Per a 9 induced human basophil activation
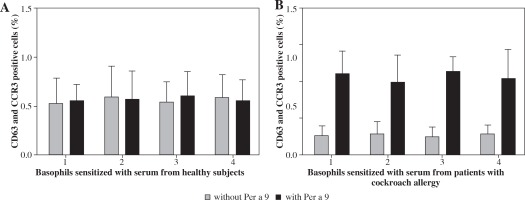
Per a 9 at 1.0 μg/ml induced approximately up to 5.6-fold increase in CD63 and CCR3 double positive cells when incubating with passively sensitized basophils by sera from the patients with American CR allergy. Per a 9 had no effect on the basophils sensitized by the sera from healthy subjects (Fig. 3).
Fig. 3
Induction of basophil activation by Per a 9. After non-specific IgEs on basophils were stripped off, cells from each donor were passively sensitized with sera from 4 different HCs (A) or from 4 different patients with American CR allergy (B), and were then challenged with Per a 9 at 1.0 μg/ml. The values shown are mean ±SE for the sera from 4 different subjects
Discussion
Numerous insect allergens such as Api m 1 [27], Api m 2 [28] from honeybee venom, Dol m 5 from bald-faced hornet (Dolichovespula maculate) [29], Sol i 3 from ant venom [30], Cul s 1 from the North American midge (Culicoides sonorensis) [31], Der f 1 from house dust mite [32], Blo t 11 from dust-mite (Blomia tropicalis) [33], Lep d 2 from dust mite (Lepidoglyphus destructor) [20], and Aed a 1 [34], Aed a 2 [35] from mosquito (Aedes aegypti) have been successfully expressed in insect cells using a baculovirus expression system. They are reported to possess similar structural and biological activities with their natural forms [36]. Since the CR is an insect, it is feasible to obtain biologically active allergens expressed in baculovirus-infected insect cells. We firstly successfully expressed the American CR allergen, Per a 5 in baculovirus-infected insect cells, proving the advantages of baculovirus-infected insect cells system in expression of American CR allergens [11]. In the present study, we succeeded in producing another biologically active and highly pure American CR allergen, Per a 9, expressed in baculovirus-infected insect cells in a relatively large amount. We found that as little as 2L of Sf-9 cell culture medium was able to produce 6 mg of Per a 9, which is enough for the functional study of Per a 9.
The antigenicities of Per a 9 were determined by ELISA, immunoblot analysis and basophile activation test. They appear to be of importance for the allergic reactions induced by CR, and have a potential application for component-based diagnosis of CR allergy. We evaluated the binding of Per a 9 to specific-IgE derived from the sera of patients with American CR allergy and the results showed that 81.3% of sera from American CR allergy patients react to Per a 9, confirming that Per a 9 is a major allergen in American CR. The basophil activation test we employed herein can represent an in vivo technique for determination of allergenicity of a given allergen. We confirmed that Per a 9 was an active allergen of CR as it is able to activate basophils which are sensitized by American CR allergic sera. In our case, the expression of the CR allergen, Per a 9, in insect cells will be helpful for CR allergy diagnosis and therapy, especially for patients showing a high reaction to this major American CR allergen.


