Introduction
Age-mediated bone loss, also called senile osteoporosis, is associated with decreased periosteal and trabecular bone formation compared to its resorption [1]. These events of bone degradation increase its fragility and also the risk of fracture; the events also lead to increased risk of death and morbidity and also deteriorates the economic state [1, 2]. The conventional therapeutic approach in the management of this age-related bone loss disorder is based on blocking bone resorption using anti-resorptive therapeutic agents such as denosumab, bisphosphonates and zoledronic acid [1, 3]. All these agents suffer the disadvantage of producing side effects such as uncommon fractures of femur bone and osteonecrosis of the jaw, limiting their use for the treatment. Hence there is urgent need of a new treatment approach for correcting this bone disorder.
In the course of developing new therapies, researchers have adapted new approaches such as promotion of osteoblastogenesis which leads to bone formation hence making up for the age-mediated loss of bone [1]. In the process of osteoblastogenesis, miRNAs have been found to play an important role in preventing the age-related bone loss [4–6]. MicroRNAs are non-coding small RNAs which can suppress the process of transcription and translation via binding directly to the target site. By binding the target sites, miRs can regulate the gene expression [7]. In case of osteoporosis, the deletion of endo-ribonuclease which is responsible for generating miRs in osteoblasts has confirmed that the Dicer generated miRNAs are necessary for promoting differentiation of osteoblasts and bone accrual in the adult mice [8]. This study suggests miRs are targets in osteoblasts and may serve as an alternative approach for correcting osteoporosis. Hence, we decided to study the pathway/mechanisms through which miRs can negatively modulate the process of bone formation and osteoblastogenesis.
In the present study microarray screening for miRs was done in bone marrow derived mesenchymal stem cells during the process of conversion from the proliferation to the mineralization state. The analysis suggested down-regulation of the number of miRs, with significant down-regulation of miR-22. In the course of the study we found that transfecting miR-22 antagomir resulted in stimulation of the pre-osteoblast process of differentiation to a phenotype which was highly differentiated and mineralized; this was associated with overexpression of COL1A1, CBFA1 and ALP proteins. We also evidenced that these effects were YWHAZ dependent. In animal studies, the treatment with miR-22 antagomir produced protective action by preventing the bone loss in ovariectomized animals and also enhanced the bone strength along with sham mice. Further in our study, the bone marrow derived stromal cells obtained from miR-22 antagomir injected mice were cultured, the results suggested increased mineralization and osteoblastogenesis, and also the levels of YWHAZ, ALP, CBFA1 and COL1A1 were upregulated; the findings were parallel to the results of in vitro studies. On this basis, miR-22 seems to exert suppressive action on bone mineralization, differentiation of osteoblasts and the process of bone formation via the YWHAZ pathway. Hence, blockade of miR-22 can be a potential approach for managing osteoporosis.
Material and methods
Collection of human bone marrow
For the study human bone marrow (BM) was collected. The volunteers were educated about the study and written informed consent was obtained from them. The protocol received approval from the ethical committee of the second affiliated hospital of Nanchang University, China; the approval number was SNU1487-1. The bone marrow aspirates were collected from five healthy, pre-menopausal female donors receiving no medication, aged 34 to 42 years. The BM was collected and processed to separate the mesenchymal stem cells by a density based fractionation system (Alpha Biotech, Glasgow, UK) and were differentiated osteogenically as described earlier [9]. For the same, the cells were cultured in α-modified Eagle’s medium (β-MEM) with added fetal bovine serum (FBS) (10%), streptomycin (0.1 mg/ml), penicillin (100 U/ml) and L-ascorbic acid-2-phosphate (0.1 M). After incubation for 4 days, the non-adhered cells were removed, considering the time point as ‘time 0’. After this the leftover adherent cells were cultured in fresh medium which contained dexamethasone (1 × 10–7 M) to prepare the bone differentiation medium; β-glycerophosphate (10 mM) was added to prepare bone mineralization media. The cells were harvested at time 0, termed as the time of proliferation, and at the time point when bone differentiation medium was added (point for differentiation); after 7 days mineralization medium was added to the medium, which was termed the point of mineralization. The viability of cells was recorded using the AlamarBlue cell viability assay. To validate the osteogenic phenotype, osteocyte-specific transcript expression, alkaline phosphatase activity and formation of mineral nodules were done. All the cell cultures showed formation of mineral nodules at the point of mineralization.
Alkaline phosphatase and alizarin red S assays
Alkaline Phosphatase Assay was done using an Alkaline phosphatase kit (BioVision, San Francisco) following the supplied instructions. Briefly, 1 × 106 cells were transferred to a 96-well plate, followed by measurement of optical density at 405 nm using a UV-visible spectrophotometer (Shimadzu 1800). The activity of alkaline phosphates was normalized against the content of DNA using a Helixyte dsDNA Quantitation Kit (AAT bioquest, California).
For Alizarin Red S assay, the cells were subjected to fixing using ice cold ethanol (70%) for 60 min. The cells were rinsed with distilled water followed by staining using Alizarin Red S (40 mM) (pH 4.2) for 10 min under continuous agitation. The cells were viewed under an optical microscope for Alizarin Red S positive cells.
The mineralization analysis was done by solubilizing Alizarin Red S staining in glacial acetic acid (10%) for 25 min and then ammonium hydroxide (10%) was added for 30 min to reach the pH of 4.2; the absorption was recorded at 405 nm using a UV-visible spectrophotometer. The extent of mineralization was normalized against DNA content using a Helixyte dsDNA Quantitation Kit (AAT bioquest, California).
Microarray analysis
Microarray analysis was done for expression of miRNA on the isolated bone marrow derived MSCs as reported earlier [9]. The microarray analysis was done to determine the alterations in expression of miRNA in various phases of differentiation. In the procedure, total RNA was isolated from the cells with the help of TRIzol reagent (Thermo Fisher, USA) at the time point of proliferation, differentiation time and mineralization time. The integrity of RNA was evaluated by electrophoresis and the content of RNA was calculated using a spectrophotometer (Shimadzu 1800). The profiling of human miRNA was done by Affymetrix microarray analysis (Thermo Fisher USA). The samples were labeled using FlashTag Biotin HSR RNA Labeling Kit (Thermo Fisher USA). The expression was compared between miRNA expression at various time points of proliferation, differentiation and point of mineralization.
Real-time TaqMan PCR miRNA analysis
In order to quantify the expression of mature has-miRNA, reverse transcriptase primers and TaqMan probes were used. For the expression GAPDH was selected as a loading control.
Quantitative real-time-PCR (qRT-PCR)
The total RNA was isolated using RNA extraction reagent (Sigma-Aldrich USA). About 2 µg of RNA was purified and subjected to reverse transcription in the cDNA along with a Prime Script 2 cDNA kit. The quantitative RT-PCR analysis was done using the Prime Script Master Mix on a CFX RT-PCR detection unit (Bio-Rad USA). GAPDH was used as a loading control.
Cultures of Iry human fetal calvarial pre-osteoblasts
The Iry human-fetal calvarial pre-osteoblast cell cultures were proliferated as discussed earlier [10]. Briefly, surgical isolation was carried out for removing individual calvaria from the skull tissues. The isolated calvaria was digested in collagenase-P (0.1%) and trypsin (0.05%) repeatedly for 20 min for releasing calvarial pre-osteoblasts. The first digestion withdrawal was discarded; from the next four digestions the cells were collected. The pre-osteoblasts were cultured in Minimum Essential Medium Eagle – α modification (Alpha MEM) without nucleosides supplemented with fetal bovine serum (10%), streptomycin (100 µg/ml) and penicillin (100 U/ml) until the population reached approximately 80% prior to further protocols.
miRNA transfection of human pre-osteoblasts in vitro)
For the study, miR-22 inhibitor (antagomir), mimic (agomir) and YWHAZ siRNA, miR-C and siCtrl served as negative controls; all were procured from Sigma-Aldrich USA. The culture medium was replaced every 6 h; the cells were collected after indicated time intervals for analysis.
Western blotting
The harvested cells were subjected to lysis in ice cold buffer (Thermo Fisher USA). The levels of proteins were measured by a protein estimation kit (Thermo Fisher USA). About 20 µg of proteins in each lane was isolated by sodium-dodecyl sulfate poly-acrylamide gel electrophoresis (12%) (SDS-PAGE) followed by transfer to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked for 60 min at 37ºC with serum albumin (bovine) (5%), followed by incubation with Iry antibodies for the next 12 h at 37ºC. The Iry antibodies used were diluted as described, Akt (1 : 1000), p-Akt (Ser-473) (1 : 1000), YWHAZ (1 : 1000), SMURF2 (1 : 1000), ALP (1 : 10000), CBFA1 (1 : 1000), COL1A1 (1 : 1000) and GAPDH (1 : 000) (Cell Signaling Tech., USA) for 60 min at 37°C; the membranes were developed using enhanced chemiluminescence reagent (Sigma-Aldrich USA). The enhanced chemiluminescence signals were collected and analyzed using a chemiluminescence imaging system (Biocompare USA).
Alkaline phosphatase (ALP) Reporter Gene Assay
293 [HEK-293] (ATCC CRL-1573) (Human embryonic kidney 293 cells) cells received transfection using miR-C, miR-22 agomir or miR-22 antagomir along with the alkaline phosphatase luciferase reporter (ALP-Luc) and cytomegalovirus promoter driven β-galactosidase reporter (pCMV-bgal). The alkaline phosphatase luciferase reporter consists of the ALP osteoblast specific regulatory element. A luciferase reporter assay kit (BioVision, San Francisco) was used for measuring luciferase activity. The β-galactosidase reporter activity was used as a control for normalizing luciferase activity. TargetScan (http://www.targetscan.org/vert_72/) along with miRanda was used in bioinformatic screening for predicting the miR-22 target site and putative targets for has-miR-22 [11].
Animal studies
For the in vivo studies, BALB/c mice (female) weighing 20 ±2 γ were selected and were housed under controlled conditions of temperature and humidity (23–24°C, 50–60% RH) under a 12 h light/dark cycle. The animal study protocols were approved by the ethical review board of the Second Affiliated Hospital of Nanchang University, Nanchang, China; the approval number was NU/301/014. The mice were given a standard chow diet along with water ad libitum. miR-C (7 mg/kg), miR-22 antagomir (7 mg/kg) or phosphate buffered saline (0.2 ml) was administered via RNAi delivery reagent Invivofectamine 3.0 Reagent (Thermo Fisher USA) into sham-operated or ovariectomized (OVX) mice for 6 weeks, on day 1 to 3 during the 1st, 3rd and the 5th week.
Micro computed tomography (MCT) analysis was done on a micro-CT scanner (SKYSCAN 1276, Bruker, USA). Briefly, the vertebrae, murine femora and tibiae were scanned at resolution of 18 mm and about 120 projections were recorded at an angle of 180°. The imaged slices were rebuilt again using a previously described algorithm [12]. The ratio of bone volume (BV) to tissue volume (TV), i.e. BV/TV, trabecular separation (Tb.Sp) and trabecular number (Tb.N) were calculated as per the procedure described earlier [13].
The formation of femoral bone was evaluated using the tetracycline labeling method via tetracycline injection (20 mg/kg) after every 30 days from the start of the protocol. Fluorescence imaging was done for calculating mineral apposition rate (MAR), mineralizing surface (MS) and bone formation rate (BFR).
For measuring the levels of ALP in vivo and the bone mineralization, the BMCs were isolated from femoral bone marrow in the differentiation medium. The BMCs were incubated and cultured for 3 weeks in bone differentiation medium prior to the assay; the culture medium was replaced with fresh medium every 48 h.
Ethics approval
The protocol received approval from the ethical committee of the Second Affiliated Hospital of Nanchang University, China, approval number SNU1487-1.
Statistical analysis
All the statistical analysis was done using GraphPad Prism software version 7. The data presented are means ± standard error of mean (SEMs) for each experiment done in triplicate. Paired t-test was done to determine differentially expressed miRs. Student’s t test was performed for comparisons. For multiple results one-way ANOVA and the Newman-Keuls test were conducted. P-values < 0.05 were considered as significant.
Results
Profiling of miRNAs in adult human bone marrow-derived MSCs transiting from proliferation-mineralization state
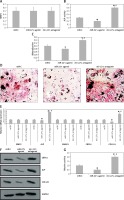
In the present study we initially evaluated the adult-human bone marrow-derived MSCs transiting from proliferation to mineralization state; it is a transition by which MSCs transform to mature osteoblasts involved in formation of bones. We observed that the cultured MSCs at the differentiation stage showed over-expressed mRNA levels of various osteoblast genes which included CBFA1, BMP2, ALP, COL1A1 and SMAD1 (p < 0.05, Figure 1 A). The outcomes of microarray analysis suggested that expression of about 10 miRs was suppressed at the differentiation time point (Figure 1 B). We further validated the suppression of all the 10 miRs using TaqMan miRNA assay (p < 0.05, Figure 1 C). Among all the observed miRNAs we found about 10-fold down-regulation in expression of miR-22 at the differentiation time point (Figure 1 C). The results of qRT-PCR for expression of miR-22 suggested that levels of miR-22 throughout the process of osteoblastic differentiation remained down-regulated significantly (p < 0.05, Figure 1 D). Looking into the findings of qRT-PCR, we selected miR-22 as a target miR for our further investigation.
Figure 1
Profiling of miRNA from proliferation to mineralization stages in human adult bone marrow-derived stem cells. A – Results of qRT-PCR for fold changes in expression of mRNA for osteoblast genes COL1A1, SMAD1, ALP, BMP2 and CBFA1 at various time points compared to proliferation time point (T0 – time 0, DT – differentiation time). B – Microarray analysis by heat map showing downregulated expression of miRs at differentiation time points compared to T0. C – Results of qRT-PCR suggesting expression of miR-22 at the T0, DT and point of mineralization. D – relative miR-22 expression
The results presented are mean ± SEM. *P < 0.05 compared to proliferation time at 0 min (T0).

miR-22 inhibits differentiation of human osteoblasts
We analyzed the effects of miR-22 antagomir or agomir transfection on the Iry human pre-osteoblasts. We observed that miR-22’s antagomir or agomir caused no significant effects on release of BMP2 (p > 0.05, Figure 2 A). However, the transfection of antagomir elevated the activity of ALP, clone formation of mineralized nodules and mineralization (Figures 2 B–D). Opposite to this, miR-22’s agomir showed its effect only on activity of mineralization and AP (Figures 2 B–D).
Figure 2
miR-22 inhibits the differentiation of osteoblasts. A–C – Effect of miR-22 antagomir or miR-22 agomir on pre-osteoblast expression levels of BMP2 (A), AMP activity (B) and mineralization (C). D – Images of Alizarin positive formation of cone in pre-osteoblast cultures. E – mRNA levels of osteoblastic genes COL1A1, SMAD1, BMP2, ALP and CBFA1. F – Western blot analysis for protein expression of COL1A1, CBFA1 and ALP. G – Effect of antagomir and agomir of miR-22 on ALPLuc and ALP promoter
Results are presented as ± SEM. *P < 0.05 compared to miR-C, #P < 0.05 compared to miR-22 agomir.
The results of qRT-PCR revealed that miR-22’s antagomir elevated the mRNA expression of COL1A1 as well as of ALP, whereas the miR-22’s agomir showed inverse effects (p < 0.05, Figure 2 E). However, the expression of protein by western blot suggested that miR-22 antagomir elevated the expression of COL1A1, CBFA1 and ALP, while the agomir showed opposite effects (Figure 2 F). These findings suggest that miR-22 antagomir modulates the expression of CBFA1 at the post-transcription level.
Earlier, a report confirmed that ALP is a favorable target gene of CBFA1 [14]. Hence we utilized ALP-Luciferase to find out whether miR-22’s antagomir stimulates the activity of ALP. The results suggested that miR-22’s antagomir stimulated the ALP promoter activity and the agomir presented a reverse effect (Figure 2 G). Altogether, the findings of the experiment suggested that miR-22 suppresses differentiation of human osteoblasts and mineralization.
miR-22 downregulates the expression of YWHAZ in human pre-osteoblasts
For prediction of miR-22’s target genes we performed bioinformatics analysis using the miRanda and TargetScan database. The database suggested that YWHAZ was the putative target gene of miR-22, which showed a match of at least 6 nucleotide sequences with miR-22 in the 3'UTR region (Figure 3 A). Parallel to this, miR-22’s agomir suppressed the expression of YWHAZ, whereas miR-22’s antagomir failed to do it (Figure 3 B). The findings were further confirmed by western blot analysis by evaluating the protein levels (Figure 3 C). A study previously confirmed that YWHAZ promotes AKT-mediated degradation of SMURF2 [15]; hence we also evaluated the role of miR-22’s antagomir and agomir in the activation of AKT and SMURF2 expression. As predicted, we evidenced that miR-22’s antagomir upregulated, whereas the agomir down-regulated the AKT induced degradation of SMURF2 (Figure 3 C).
Figure 3
miR-22 suppresses expression of YWHAZ in human pre-osteoblasts. A – Bioinformatic analysis of binding of miR-22 to YWHAZ’s 3′UTR region. B – mRNA expression of YWHAZ by qRT-PCR in pre-osteoblasts after 24 h of transfection with miR-22 antagomir and miR-22 agomir. * P < 0.05 compared to miR-C, #p < 0.05 compared to miR-22 agomir. C – Western blot analysis for protein expression of SMURF2, YWHAZ, p-AKT and AKT in pre-osteoblasts after 24 h of transfection with miR-22 agomir or miR-22 antagomir. D – Protein expression of YWHAZ in pre-osteoblasts transfected with siRNA or si-control compared to siYWHAZ or siControl. E–G – BMP2 secretion, AMP activity and mineralization in osteoblasts transfected with siControl or siYWHAZ. H – mRNA expression of osteoblast genes COL1A1, SMAD1, BMP2, ALP and CBFA1 by qRT-PCR. I – Western blot analysis of AKT, p-AKT, COL1A1, SMURF2, ALP and CBFA1 in osteoblasts transfected with siYWHAZ or siControl

After confirming that miR-22 suppresses the expression of YWHAZ, we evaluated the effect of YWHAZ blockade on the process of osteoblast differentiation. We observed that inhibition of YWHAZ by siRNA (Figure 3 D) did not impact the release of BMP2 (Figure 3 E) but significantly decreased the ALP activity, mineralized nodule clone formation and mineralization (Figures 3 F, G). Further, the results of qRT-PCR suggested that YWHAZ blockade decreased the mRNA expression of COL1A1 and ALP significantly (p < 0.05, Figure 3 H). However, the results of western blot analysis suggested that the blockade of YWHAZ decreased the expression of COL1A1, CBFA1 and ALP significantly (p < 0.05, Figure 3 I). The results confirmed that blockade of YWHAZ at the post-transcription level modulates the expression of CBFA1.
A previous report suggested that AKT-induced degradation of SMURF2 promotes CBFA1 stability [15]; hence we evaluated the effect of YWHAZ blockade on protein expression of SMURF2 and activation of AKT. As expected, the results showed that blockade of YWHAZ suppressed the AKT-induced degradation of SMURF2 and upregulated the levels of SMURF2 (Figure 3 I). It is already established that SMURF2 causes degradation of CBFA1 protein, and the over-expressed SMURF2 leads to decreased levels of CBFA1 protein (unaltered CBFA1 mRNA levels) in si-TWHAZ cells (Figures 3 H, I). All together, miR-22 suppresses the YWHAZ/AKT induced stability of CBFA1 protein via enhancing the SMURF2 associated degradation of CBFA1, hence inhibiting differentiation of osteoblasts.
miR-22’s antagomir reverses the ovariectomized-mediated bone loss and attenuates bone strength of experimental mice
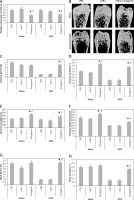
In order to analyze the anabolic effect of miR-22’s antagomir in an animal model, the ovariectomized or the sham operated mice were injected with phosphate buffer saline, miR-C and antagomir for a period of 6 weeks. At first, we analyzed the expression miR-22 in the bone extracts to confirm the delivery of miR constructs to the bone tissues (p < 0.05, Figure 4 A). The study also evaluated the trabecular bone at the tibial metaphysis, the femoral metaphysis and the vertebral column (Figure 4 B). The ovariectomized mice showed significant trabecular loss as demonstrated by decreased values of BV/TV, Tb.N and elevated Tb.Sp in all the 3 bone regions (p < 0.05, Figure 4 C, D and G–L). We observed that these damaging effects in ovariectomized mice were partially rescued by miR-22’s antagomir (p < 0.05), whereas the treatment of miR-C and PBS showed no significant changes. In addition, miR-22’s antagomir enhanced the stiffness and strength of femoral bone of ovariectomized as well as sham operated mice (p < 0.05, Figures 4 E, F). Parallel to the results of miR-22’s antagomir mediated increase in Tb.N (Figure 4 D), a significant increase in levels of BFR and MAR was observed; both of these are important femoral bone formation parameters (Figures 4 N, O). All together, the experiment confirmed that miR-22’s antagomir reversed the ovariectomized mediated bone loss and also attenuated the bone strength in vivo.
Figure 4
miR-22 antagomir reverses the bone loss in ovariectomized mice and attenuates bone strength. A – decreased expression of miR-22 in bone after injection of miR-22 antagomir in the Balb/c mice, B – micro-CT image of femoral bone in ovariectomized and sham operated mice, C–F – data of micro-CT parameters which include BV/TV ratio, Tb.N, Tb.Sp, bone strength and stiffness, G–H – epiphyses of tibia I – epiphyses of tibia, J–L – vertebrae, M – the mineralizing surface, N – the mineral apposition rate and bone formation rate, O – bone formation rate
The results are presented as mean ± SEM. *P < 0.05 compared to mice PBS treated. #P < 0.05 compared to miR-C group.

miR-22’s antagomir inhibits the levels of YWHAZ in vivo and stimulates osteoblasts in mice
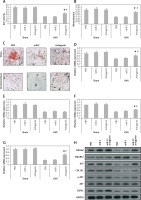
In order to explore the involved mechanism responsible for the anabolic effect of antagomir on bones, the stromal cells were separated from the bone marrow of mice at the end of the 6th week. As expected, the ovariectomized mice showed decreased ALP activity, formation of mineralized nodules and mineralization from the bone marrow isolated stromal cells (p < 0.05, Figures 5 A–C). The results also showed that the damaging effect in ovariectomized mice was reversed to some extent by miR-22’s antagomir, whereas the treatment of miR-C and phosphate buffered saline failed to show any significant effects. Consistently with our in vitro results (Figure 3), the bone-marrow derived stromal cells isolated from mice treated with antagomir showed significant mRNA overexpression of ALP activity along with expression of COL1A1 and YWHAZ (Figures 5 D–G), whereas the results of western blot analysis demonstrated significant overexpression of YWHAZ, COL1A1, ALP and CBFA1 (Figure 5 H). All together, the effects of miR-22’s antagomir are mediated via upregulation of YWHAZ and activation of ALP.
Figure 5
miR-22’s antagomir inhibits in vivo YWHAZ mediated expression by stimulating osteoblastogenesis. The bone marrow derived stromal cells were harvested from sham mice or from the ovariectomized mice treated with phosphate buffered saline, miR-C or with miR-22’s antagomir. A – ALP activity, B – mineralization, C – colony formation of alizarin positive cells, D–G – the qRT-PCR analysis for mRNA levels of YWHAZ (D) CBFA1 (E) ALP (F) and COL1A1 (G), H – Western blot analysis for expression of YWHAZ, p-AKT, AKT, CBFA1, SMURF2 and ALP
The results are presented as mean ± SEM. *P < 0.05 compared to phosphate buffered saline treated group. #P < 0.05 compared to miR-C treated group.
Discussion
MicroRNAs are regulators of a number of processes by regulating the translation of mRNA [7]. Due to the damaging and weakening consequences of senile osteoporosis, there is an urgent and increasing need for decoding the roles of miRNA in the process of bone development, remodeling, growth and homeostasis [4–6]. To date numbers of miRNAs have been identified to be linked with these processes. Some miRNAs are identified as promoters of osteoblast differentiation; e.g. miR-29b is reported to increase differentiation of osteoblasts in pre-osteoblasts via suppressing the inhibitors of osteogenesis; the target proteins involved were HDAC4, DUSP2, TGFβ3, AcvR2b, and CTNNIBP1 [15]. miR-29c promotes Wnt signaling in osteoblasts by targeting inhibitors, and regulates matrix proteins; the target proteins of miR29c were Dkk1, Kremen, SFRP2, and osteonectin [16]. miR-196a targets Hoxc8 and its overexpression in adipose derived MSCs leads to differentiation of osteoblasts [17]. miR-378 targets nephronectin, an extracellular protein which competes for binding of miR and promotes differentiation of osteoblasts [18]. Some miRNAs have also been found to inhibit the process of differentiation in osteoblasts; e.g. miR-637 has been found to promote adipocytes and inhibits differentiation of osteoblasts via osterix [19]. Cluster of miR-23a-27a-24-2 exhibits inhibitory action on the differentiation of osteoblasts via downregulation of RUNX2 protein, hence promoting osteogenesis [20].
In the present study, the expression of miR in adult bone marrow derived stem cells suggested significant downregulation of miR-22 during the very important period of proliferation-to-mineralization transition [21]. MiRNA-22 is an important multifunctional miRNA; it is found to inhibit proliferation of osteosarcoma cells and also blocks the migration by targeting HMGB1 and related autophagy [22]. miR-22 is also found be downregulated in hepatocellular carcinoma and exerts an antiproliferative effect on cancer cells both in vitro and in vivo [23]. This evidence suggested that miR-22 can play an important role in suppressing osteoblastogenesis.
In the present work we used antagomir; the results demonstrated that miR-22 promoted the differentiation of pre-osteoblasts into an improved phenotype which was more mineralized and differentiated. We also found that the differentiation was YWHAZ dependent, and the phenotypes showed upregulated protein expression of COL1A1, ALP and CBFA1. In a previous study, Chen et al. evidenced that YWHAZ was a favorable target of miR-22 for inhibiting metastasis of hepatic cancer. The study by Chen et al. also suggested that miR-22 inhibited YWHAZ mediated phosphorylation of AKT in hepatic carcinoma cells [24]. However, it has been found that YWHAZ is responsible for activating AKT signaling, which in turn elevates the protein stability of CBFA1 by decreasing SMURF2 mediated degradation of CBFA1 [15]. However, it is also confirmed that CBFA1 is necessary for mesenchymal stem cell associated osteoblastic cell lines [25]. Hence the strategy for inhibiting miR-22 is to target the protein (SMURF2) which is responsible for degradation of CBFA1, though it is found that agents inhibiting the protein, i.e. SMURF2, are not yet developed; developing such agents would be an interesting approach for promoting the differentiation of osteoblasts in subjects suffering from osteoporosis.
In our study we found that miR-22 did not alter the expression of SMAD1 and BMP2 in vitro. As it is reported that both SMAD1 and BMP2 are the upstream targets of CBFA1 and AKT [13, 15, 26], the expression levels of SMAD1 and BMP2 were not altered by miR-22 mediated regulation of YWHAZ/AKT-induced stability of CBFA1 protein.
The results of in vivo study suggested that anti-miR-22 prevented the trabecular bone loss in experimental ovariectomized mice and also improved the bone strength in sham operated as well as in ovariectomized mice. Hence the outcomes of our study would suggest focusing on inhibition of miR-22 as a therapeutic approach for promoting osteoblastogenesis in osteoporotic subjects. Though presence of agents, such as AMO-22, which act as inhibitors of miR-22 can be utilized [27], direct inhibition via synthetic anti-miR-22 antagomirs or high throughput screening may be strategies for inhibiting miR-22.
In conclusion, miR-22 blocks osteoblastogenesis in both in vitro and in vivo models and may serve as a potential therapeutic approach in treatment of osteoporosis clinically.



