Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
ONCOLOGY / BASIC RESEARCH
Long intergenic non-coding RNA, regulator of reprogramming (LINC-ROR) over-expression predicts poor prognosis in renal cell carcinoma
1
Department of Medical Biochemistry and Molecular Biology, Faculty of Medicine, Suez Canal University, Ismailia, Egypt
2
Department of Biochemistry, Faculty of Medicine, Northern Border University, Arar, Saudi Arabia
3
Genetics Unit, Department of Histology and Cell Biology, Faculty of Medicine, Suez Canal University, Ismailia, Egypt
4
Center of Excellence of Molecular and Cellular Medicine, Suez Canal University, Ismailia, Egypt
5
Botany Department, Faculty of Science, Suez Canal University, Ismailia, Egypt
Submission date: 2018-04-19
Final revision date: 2018-06-06
Acceptance date: 2018-07-14
Online publication date: 2019-05-17
Publication date: 2021-07-16
Arch Med Sci 2021;17(4):1016-1027
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Long intergenic non-coding RNA, regulator of reprogramming (LINC-ROR) is a newly identified cytoplasmic long non-coding RNA (lncRNA) implicated in cell longevity and apoptosis. We aimed in the current work for the first time to investigate the association of the expression profiles of LINC-ROR and three stem-related transcriptional factors with clinicopathological data and their impact on renal cell carcinoma (RCC) progression in a sample of RCC patients.
Material and methods:
Expression levels of LINC-ROR and stemness-related factors: SOX2, NANOG, and POU5F1 were detected in 60 formalin-fixed, paraffin-embedded tissues, and their paired adjacent non-cancer tissues (n = 60) by using real-time qRT-PCR analysis. Additionally, the expression profiles were compared with the available clinicopathological features.
Results:
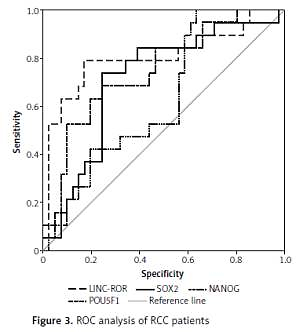
The genes studied were markedly up-regulated in RCC (medians and interquartile ranges were 30.3 (1.84–235.5), 10.2 (1.84–53.9), 5.39 (0.94–23.5), and 12.5 (1.61–43.2) for LINC-ROR, SOX2, NANOG, and POU5F1, respectively) relative to paired non-cancer tissue. High expression levels were associated with poor prognosis in terms of tumour undifferentiation (for LINC-ROR, SOX2, and NANOG), lymph node infiltration (for SOX2), postoperative recurrence (for LINC-ROR and SOX2), and shorter overall survival (OS) and progression-free survival (for all genes studied). The best curve for OS prediction was constructed with LINC-ROR data (area under the receiver operating characteristic curve (AUC) = 0.804 at a cut-off value of 72.7, sensitivity 78.9%, and specificity 80.5%).
Conclusions:
Collectively, aberrant LINC-ROR and pluripotent gene expression may be recognised as prognostic markers for RCC. Future functional studies are highly recommended to validate the study findings.
Long intergenic non-coding RNA, regulator of reprogramming (LINC-ROR) is a newly identified cytoplasmic long non-coding RNA (lncRNA) implicated in cell longevity and apoptosis. We aimed in the current work for the first time to investigate the association of the expression profiles of LINC-ROR and three stem-related transcriptional factors with clinicopathological data and their impact on renal cell carcinoma (RCC) progression in a sample of RCC patients.
Material and methods:
Expression levels of LINC-ROR and stemness-related factors: SOX2, NANOG, and POU5F1 were detected in 60 formalin-fixed, paraffin-embedded tissues, and their paired adjacent non-cancer tissues (n = 60) by using real-time qRT-PCR analysis. Additionally, the expression profiles were compared with the available clinicopathological features.
Results:
The genes studied were markedly up-regulated in RCC (medians and interquartile ranges were 30.3 (1.84–235.5), 10.2 (1.84–53.9), 5.39 (0.94–23.5), and 12.5 (1.61–43.2) for LINC-ROR, SOX2, NANOG, and POU5F1, respectively) relative to paired non-cancer tissue. High expression levels were associated with poor prognosis in terms of tumour undifferentiation (for LINC-ROR, SOX2, and NANOG), lymph node infiltration (for SOX2), postoperative recurrence (for LINC-ROR and SOX2), and shorter overall survival (OS) and progression-free survival (for all genes studied). The best curve for OS prediction was constructed with LINC-ROR data (area under the receiver operating characteristic curve (AUC) = 0.804 at a cut-off value of 72.7, sensitivity 78.9%, and specificity 80.5%).
Conclusions:
Collectively, aberrant LINC-ROR and pluripotent gene expression may be recognised as prognostic markers for RCC. Future functional studies are highly recommended to validate the study findings.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.