Introduction
Glioblastoma multiforme (GBM) is a kind of malignant brain tumor prevalent in adults, with the characteristics well adapted to poorly immunogenic and hypoxic conditions [1]. Effective treatment of GBM is impeded due to the high proliferation, migration and invasion of GBM cells [2]. GBM cells migrate by degrading the extracellular matrix, so it is difficult to have GBM cells eradicated completely by surgery [3]. According to the existing research, the median survival time of patients suffering from GBM is no more than 15 months, and 5-year survival prognosis of the patients is less than 10%, even if treated with aggressive therapy strategies [4, 5]. The exact molecular mechanisms of GBM cell proliferation are still unclear, so further studies on the molecular mechanisms are urgently needed for the development of effective therapeutic strategies.
MicroRNAs (miRNAs) are non-coding RNA molecules comprising 20 to 25 nucleotides [6]. MiRNAs regulate gene expression, which is associated with physiological and pathological processes [7]. Particularly, they are involved in tumorigenesis [8]. MiRNAs have been proved to have a critical relationship with various kinds of human cancers, such as lung and hepatic cancers [9, 10]. They can act as tumor suppressors by targeting oncogenes. Enhanced evidence has accumulated revealing that several miRNAs are involved in the biological processes associated with GBM progression, including cell proliferation, migration, invasion, apoptosis, etc. For example, miR-522 has been reported to be positively correlated with GBM cell proliferation by down-regulating the expression of PHLPP1 [11]. MiR-1908 has been proved to function as a GBM oncogene by suppressing the PTEN tumor suppressor pathway [12]. The investigation of miRNAs would be of great importance and the exploration of the potential miRNAs may help us identify biomarkers, as well as explore novel therapies for patients with GBM. Initially, miR-431 dysregulation was specific to the nervous system, such as spinal muscular atrophy [13]. Additionally, the down-regulation of miR-431 inhibits the viability of GBM cells by regulating SOCS6 [14]. Recent evidence has shown its relationship with development of other tumors. For instance, miR-431 was detected to have elevated expression in colorectal cancer [15], and its overexpression in hepatocellular carcinoma cells was confirmed to inhibit cell invasion and migration [16].
The dysregulation of miR-431 could affect tumorigenesis by regulating its target mRNAs. In this study, we focused on targeting the relationship between miR-431-5p and EPB41L1 in GBM cells. EPB41 encodes a multifunctional protein that mediated the communication between the erythrocyte cytoskeleton and the overlying plasma membrane. EPB41 binds to and stabilizes dopamine receptors at the neuronal plasma membrane. EPB41L1 was reported to act as a potential tumor suppressor [17]. But there is no evidence clarifying the correlation between miR-431 and EPB41L1. Mechanically, we have found that miR-431-5p enhances the proliferation and invasion of GBM cells by suppressing the expression of EPB41L1.
Material and methods
Tissue samples
Glioblastoma multiforme tissues and the adjacent tissues (located at least 2 cm from the tumor edges) were obtained from 30 patients who received surgical treatment in Cangzhou Central Hospital, Cangzhou, Hebei, China from January, 2015 to March 2016. Samples were immediately frozen in liquid nitrogen until analysis [18, 19].
Cell culture
The normal brain glial cell line HEB and GBM cell lines U251, A172, U-118 and U87 (purchased from Chinese Academy of Sciences, Shanghai, China) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) [19–22] with 10% fetal bovine serum (FBS) at 37°C under the atmosphere of 5% CO2 and 95% air.
Cell transfection
U87 cells were respectively transfected with miR-431-5p mimics (miR-mimics), miR-431-5p inhibitor (anti-miR), miR-431-5p inhibitor negative control (anti-NC), and miR-431-5p mimics negative control (miR-NC) using Lipofectamine 2000 reagent (GenePharma Co., Ltd., Shanghai, China). The lentiviral vector (plenti-GIII-UbC) was used to construct a recombinant vector (plenti-GIII-Ubc-EPB41L1) which was then transfected into U87 cells (plenti-EPB41L1). Meanwhile, U87 cells transfected with pLenti-GIII-UbC-Null (plenti-Null) were the negative control group.
RNA extraction and qRT-PCR
Frozen tissues and cell samples were defrosted, and then TRIzol reagent (Invitrogen, Carlsbad, CA) was added to extract total RNA according to the instructions. QuantiTect SYBR Green RT-PCR kit (Qiagen, Hilden, Germany) was applied to evaluate the relative expression of miR-431-5p and EPB41L1 to U6 or GAPDH (the normalizer), respectively. The expression level was also calculated using the 2–ΔΔCt method. The primer sequences (synthesized by Ribo Bio, Guangzhou, China) are given in Table I.
Western blot
Concentrations of the protein extracted from cells and tissues were measured using a bicinchoninic acid (BCA) protein assay kit (Beyotime Biotech, Haimen, China). A total of 30 μg of protein was resolved by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to immunoblot polyvinylidene difluoride membranes (Chemicon International, Millipore, Billerica, MA, USA). The membranes were blocked with 5% skimmed milk in Tris-buffered saline with 0.1% Tween (TBS-T) for 1 h, washed three times with TBS-T, and incubated for 10 h at 4°C with primary antibodies against EPB41L1 (1 : 2000) and GAPDH (1 : 2000) (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). The blots were then incubated with horseradish peroxidase-labeled secondary goat anti-rabbit (1 : 2000) or rabbit anti-goat antibodies (1 : 2000) (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). Membranes were visualized with enhanced chemiluminescence (ECL kit; Applygen Inst. Biotech, Beijing, China).
Dual luciferase reporter gene assay
The wild type or mutated 3′-UTR (replacing GCAAGAC by CGUUCG according to the prediction of TargetScan software) of EPB4L1 was inserted into the pMIR-REPORT luciferase vector (Promega, Madison, USA) to construct the pMIR-REPORT-wt or pMIR-REPORT-mut vector. The wild-type or mutated plasmid was co-transfected into U87 cells with miR-mimics or miR-NC. The empty pmirGLO vector (pGLO-NULL) was also transfected as a control. Cells were collected 48 h after transfection, and then the Dual Luciferase Reporter Assay System (Promega, Madison, USA) was used to detect the relative luciferase activity according to the protocol.
MTT assay
Fresh nutrient media were added to U87 cells after centrifugation. Then the cells (1 × 104/well) were transferred into 96-well plates. Samples were treated with MTT solution (6 mg/ml) strictly following the protocols for 4 h. After dissolving the crystals with 100 μl of DMSO, the optical density (OD) value was measured at 490 nm.
Flow cytometry analysis
Cells were fixed with 90% ethanol and then incubated with 50 mg/l RNase. Nuclei of cells were stained with 50 mg/l propidium iodide (PI) for 30 min. The cell cycle was examined using a flow-cytometer (CapitalBio, Beijing, China). Cell apoptosis was detected using the flow cytometer after incubating with Annexin V-FITC/PI reagent for 15 min.
Transwell assay
The Transwell invasion assay was performed using Matrigel invasion chambers (Corning Inc., USA). 1 × 105 cells in 15 μl medium were seeded in the upper chambers, and DMEM supplemented with 10% FBS was added to the bottom chamber to stimulate cell invasion. After 24 h of incubation, the bottom cells were fixed with 70% ethanol for 15 min, washed with PBS and stained with crystal violet. Six random fields of every well were photographed. The cell number of every view was counted under a microscope. The OD value of every well at 570 nm was measured as well.
Wound healing assay
Cells (2.5 × 105/ml) were seeded in 6-well plates and a 100 μl pipette tip was used to manually make a scratch 24 h after transfection on the surface of cell culture of each well. The scratch widths were initially measured (0 h) and re-measured after 24 h. The percentage of the shortened scratch width was defined as the wound healing rate (setting the wound healing rate as 0% initially).
Results
MiR-431-5p expression in GBM tissues and cell lines
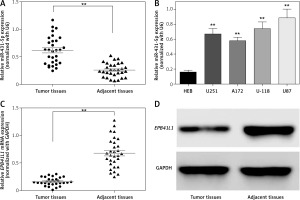
The expression of miR-431-5p was detected by qRT-PCR. We found that miR-431-5p was significantly overexpressed in GBM tissues compared to adjacent tissues (Figure 1 A, p < 0.01). MiR-431-5p was significantly overexpressed in GBM cell lines including U251, A172, U-118 and U87 compared to the normal brain glial cell line HEB (Figure 1 B, p < 0.01). The highest expression level of miR-431-5p was seen in the U87 cell line, which was selected for the following assays.
Figure 1
MiR-431-5p and EPB41L1 expression levels in GBM cells and tissues. A – Expression levels of miR-431-5p in GBM and adjacent tissues detected by qRT-PCR. B – miR-431-5p expression in different GBM cell lines and normal brain cell line HEB. C – mRNA levels of EPB41L1 in GBM and adjacent tissues detected by qRT-PCR. D – EPB41L1 protein levels in GBM and adjacent tissues detected by Western blot
GBM – glioblastoma multiforme; **p < 0.01 was considered statistically significant.
EPB41L1 was under-expressed in GBM tissues
Results of qRT-PCR showed that EPB1L1 mRNA expression was dramatically lower in GBM tissues than in adjacent tissues (Figure 1 C, p < 0.01). Similarly, the protein level of EPB1L1 detected by Western blot was also lower in GBM tissues compared with that in adjacent normal tissues (Figure 1 D).
MiR-431-5p directly targeted EPB41L1
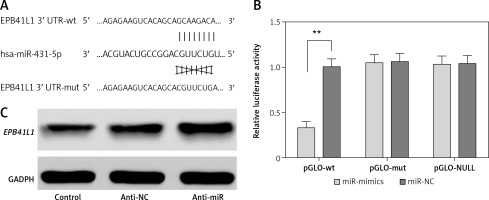
The predicted gene sequences of wild-type and mutated miR-431-5p binding sites of EPB41L1 3′UTR are illustrated in Figure 2 A. PGLO-wt and pGLO-mut were constructed carrying wild-type or mutated EPB41L1 3′UTR. The results of dual luciferase reporter gene assay indicated that with the overexpression of miR-431-5p, the relative luciferase activity in the pGLO-wt group dramatically decreased compared with that in the miR-NC group (Figure 2 B, p < 0.01), while the pGLO-mut group showed no significant difference between pGLO-wt and miR-NC groups. The results of Western blot showed that the inhibition of miR-431-5p upregulated the expression of EPB41L1 in U87 cells (Figure 2 C). In conclusion, miR-431-5p could directly target EPB41L1 and inhibit its expression in GBM cells.
Figure 2
MiR-431-5p directly targeted EPB41L1 and inhibited its expression. A – Gene sequences of wild-type and mutated miR-431-5p binding sites of EPB41L1 3′UTR predicted by TargetScan. B – Luciferase activity of each group detected by dual luciferase reporter gene system. C – EPB41L1 expression in transfected U87 cells detected by Western blot
**p < 0.01 was considered statistically significant.
Influence of miR-431-5p and EPB41L1 expression on proliferation of GBM cells
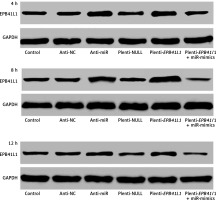
Expression of miR-431-5p and EPB41L1 in 4, 8 and 12 h
Figure 3 shows the of EPB41L1 expression 4, 8 and 12 h after transfection in different groups. It was found that EPB41L1 expression significantly increased in the anti-miR group and the plenti-EPB41L1 group throughout the 12 h after transfection. However, the restoration experiments showed that within 4 h after the transfection, the inhibitory effect of miR-mimics on the expression of EPB41L1 was the strongest. After 4 h of transfection, the inhibitory effect of miR-mimics on the expression of EPB41L1 decreased but remained steady.
Figure 3
Expression of miR-431-5p and EPB41L1 in 4, 8 and 12 h after transfection in different groups. It was shown that EPB41L1 expression significantly increased in the anti-miR group and the plenti-EPB41L1 group throughout the 12 h after transfection. Within 4 h after transfection, the inhibitory effect of miR-mimics on the expression of EPB41L1 was the strongest. After 4 h transfection, the inhibition of miR mimics on the expression of EPB41L1 decreased but remained steady
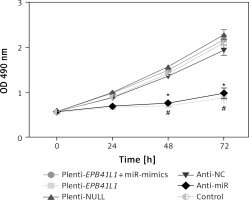
Proliferation of GBM cells detected by MTT assay
As shown in MTT assay results (Figure 4), the growth rate of U87 cells after transfection of miR-431-5p inhibitors significantly decreased compared with that in the control group (p < 0.01). Meanwhile, U87 cells transfected with plenti-EPB41L1 had a lower growth rate than those in the plenti-NULL group (p < 0.01), while U87 cells transfected with anti-NC or plenti-NULL, and cells co-transfected with plenti-EPB41L1 and miR-mimics showed no significant difference compared with the control group. The above results indicated that down-regulated expression of miR-431-5p and over-expressed EPB41L1 both inhibited the proliferation of GBM cells.
Figure 4
Influence of miR-431-5p and EPB41L1 expression on proliferation of GBM cells. Proliferation of U87 cells in different groups detected by MTT assay
GBM – glioblastoma multiforme; *p < 0.05 compared with anti-NC group corresponding timing point, #p < 0.05 compared with plenti- NULL group corresponding timing point.
Influence of miR-431-5p and EPB41L1 expression on GBM cell cycle progression and apoptosis
Cell cycle progression detected by flow cytometry
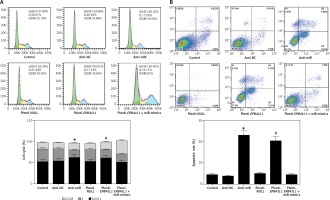
As shown in Figure 5 A, both in the anti-miR group and the plenti-EPB41L1 group, the percentage of U87 cells in S phase was significantly lower than that in the control group (p < 0.01), while that of cells in G0/G1 phase was significantly higher (p < 0.05). However, the percentage of cells in each phase of anti-NC, plenti-NULL and plenti- EPB41L1 + miR-mimic groups did not show a statistically significant difference compared with the control group. Therefore, it was proved that decreasing miR-431-5p expression and increasing EPB41L1 expression both arrested cell growth in G0/G1 stage and inhibited cell proliferation, which could be hindered by miR-431-5p overexpression.
Figure 5
Influence of miR-431-5p and EPB41L1 expression on apoptosis of GBM cells. A – Cell cycle progression of U87 cells 48 h after transfection in different groups detected by flow cytometry; B – cell apoptosis of U87 cells in different groups 48 h after transfection detected by flow cytometry. Cell apoptosis rate in anti-miR and plenti-EPB41L1 groups was compared with the control group
GBM – glioblastoma multiforme; *p < 0.05 compared with anti-miR group, #p < 0.05 compared with plenti-NULL group.
Cell apoptosis detected by flow cytometry
Cells in anti-miR and plenti-EPB41L1 groups showed a significantly higher apoptosis rate compared with those in the control group (Figure 5 B, p < 0.001), while there was no significant difference between the control group, anti-NC, plenti-NULL and plenti-EPB41L1 + miR-mimics groups. Therefore, it was demonstrated that decreasing miR-431-5p expression and increasing EPB41L1 expression both promoted cell apoptosis.
Influence of miR-431-5p and EPB41L1 expression on invasion and migration of GBM cells
Invasion of GBM cells detected by Transwell assay
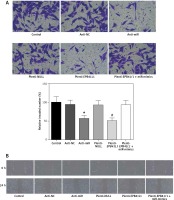
In anti-miR and plenti-EPB41L1 groups, the number of U87 cells invaded through the membrane was significantly smaller than that in the control group (Figure 6 A, p < 0.01), but there was no significant difference between control, anti-NC, plenti-NULL and plenti-EPB41L1+miR-mimics groups. In summary, it was proved that down-regulating miR-431-5p expression or up-regulating EPB41L1 expression could both reduce cell invasiveness.
Figure 6
Influence of miR-431-5p and EPB41L1 expression on GBM cell invasion and migration. A – Cell invasiveness of U87 cells in different groups detected by Transwell assay; cell invasiveness in anti-miR and plenti-EPB41L1 groups was compared with that in control group. B – Cell migration of U87 cells in different groups detected by wound healing assay. Compared with anti-miR group
GBM – glioblastoma multiforme; *p < 0.05 compared with anti-miR group, #p < 0.05 compared with plenti-NULL group.
Cell migration detected by wound healing assay
The results of the wound healing assay showed that in both anti-miR and plenti-EPB41L1 groups, the scratch width was significantly wider than that in the control group (Figure 6 B). The results indicated that decreasing the expression of miR-431-5p and increasing the expression of EPB41L1 could weaken cell migration.
Discussion
Glioblastoma multiforme is widely accepted to be one of the most common malignant tumors of the primary brain in adults [23–26]. Due to the high-level invasiveness, the prognosis is always bad with a high recurrence rate [23]. Therefore, it is of high importance to research the pathogenesis of GBM. Here, in this study, we revealed one of the possible molecular mechanisms whereby miR-431-5p may enhance the invasion and proliferation of GBM cells via suppressing EPB41L1. Through dual luciferase and Western blot assays, we have confirmed this target regulating relationship between GBM and EPB41L1. It is the first time that the target relationship between miR-431-5p and EPB41L1 in GBM cells has been studied.
Many previous studies have come to a conclusion that miRNAs act as key factors in the pathogenesis of many human cancers including gastric cancer [27], ovarian cancer [28], esophageal squamous cancer [29], bladder cancer [30], etc., and they have been considered as one of the most important factors in studies that focus on cancer pathogenesis. As for GBM, many miRNAs seem to be related to its carcinogenesis, such as miR-153 [31], miR-7-1-3p [32], miR-124 [33], and they serve as either tumor suppressors or oncomirs. We herein discovered that miR-431-5p was deregulated in GBM tissues and cells, indicating that it might be associated with GBM carcinogenesis. MiR-431 has been reported to be related to many human diseases including hepatocellular carcinoma [16, 34], spinal muscular atrophy [13] and medulloblastomas [14], etc. As for GBM, Tanaka et al. found that the down-regulation of miR-431 inhibited the viability of GBM cells [14], which is similar to our conclusion that the up-regulation of miR-431-5p could promote the viability of GBM cells. We thus believe that miR-431-5p served as an oncomir in GBM. However, in their research, miR-431 was only regarded as a down-stream target and did not focus on the down-stream target of miR-431. Many previous studies have proved that miRNAs might be mediators and target oncogenes to exert their functions [35–37]. Here in our study, we revealed that miR-431-5p could target tumor suppressors and augment the aggressiveness of GBM.
EPB41L1, also known as 4.1N, is a kind of human protein which has been defined to be closely related to ovarian cancer [38, 39] and carcinogenesis in prostate [40]. Also, Wang et al. claimed that EPB41L1 could link PP1 to JNK-c-Jun pathway regulation and this procedure implied that EPB41L1 might act as a potential tumor suppressor in non-small cell lung cancer [17]. Xi et al. [39] took epithelial ovarian cancer as an example and proved that EPB41L1 expression level significantly decreased during the malignant transformation of epithelial ovarian tumors, which can also support our results. Zhang et al. confirmed a similar relationship in ovarian cancer that EPB41L1 could suppress hypoxia-induced epithelial-mesenchymal transition in epithelial ovarian cancer cells [38]. Of all the studies on EPB41L1, we found that EPB41L1 primarily suppressed the proliferation, invasion and transition of cancer cells. And here we reported that the up-regulation of EPB41L1 might suppress the viability of GBM cells.
However, although we confirmed that miR-431-5p could target EPB41L1, and it is the first miRNA that was found to target EPB41L1, we have certain limitations in this study. First, miR-431-5p is not the only regulating factor of EPB41L1. It could be regulated by various factors including calmodulin such as Ca2+/CaM, the K+/Cl– cotransporter KCC2, dopamine receptors, and glutamate receptors ionotropic (AMPA) receptors [41–45] in a variety of cancers. Second, EPB41L1 is associated with malignancy [39, 46], and miR-431-5p is associated with malignancy [15, 16, 34], but whether the degree of the GBM malignancy has an influence on the effects of miR-431-5p or EPB41L1 needs further confirmation. Thirdly, the target relationship between miR-431-5p and EPB41L1 was found, but we could not exclude the possibility that it is a part of a signaling pathway in the whole pathogenesis of GBM. Previous studies have indicated that a miR-protein regulating relationship might be a part of a signaling pathway regulation [47–49]. Thirdly, the deviations caused by age, gender and different periods of the diseases were not included. Further research is still warranted to comprehend the pathogenesis of GBM.
In conclusion, for the first time we reported that miR-431-5p promoted the proliferation and invasion of GBM cells via suppressing EPB41L1 and we have proved it via multiple experiments including Western blot, dual luciferase reporter gene assay, flow cytometry, etc. Our research may provide useful information on the treatment against GBM, and therefore improve the prognosis of the patients.


