Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
BASIC RESEARCH
MiR-345-5p inhibits tumorigenesis of papillary thyroid carcinoma by targeting SETD7
1
Department of Head and Neck Surgery, Zhejiang Cancer Hospital, Zhejiang, China
Submission date: 2018-11-27
Final revision date: 2018-12-24
Acceptance date: 2019-01-08
Online publication date: 2019-03-25
Publication date: 2020-05-26
Arch Med Sci 2020;16(4):888-897
KEYWORDS
TOPICS
ABSTRACT
Introduction:
This study aimed to explore the effects of miR-345-5p on papillary thyroid carcinoma (PTC) and uncover the possible mechanisms.
Material and methods:
MiR-345-5p and SETD7 mRNA levels were analyzed by quantitative real-time PCR and SETD7 protein level was measured by Western blot. The viability, colony formation ability and apoptosis of PTC cells were measured with CCK-8, soft agar colony formation and flow cytometry assay, respectively. Luciferase reporter assay was used to identify miR-345-5p’s target.
Results:
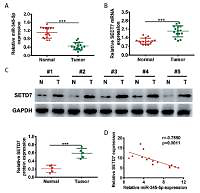
Compared to neighboring normal tissues, there was lower miR-345-5p expression and higher SETD7 expression in PTC tissues. Moreover, Spearman’s correlation analysis indicated that there was a negative correlation between miR-345-5p and SETD7 expression in PTC tissues. MiR-345-5p mimics inhibited the viability and colony formation of TPC1 and B-CPAP cells and promoted apoptosis, whereas anti-miR-345-5p promoted PTC cell proliferation and inhibited apoptosis. SETD7 was confirmed to be a direct target of miR-345-5p through target scan analysis and luciferase reporter assay. Additionally, overexpression of SETD7 promoted the viability and colony formation of TPC1 and B-CPAP cells and inhibited apoptosis, whereas downregulation of SETD7 by shRNAs had opposite effects on PTC cells. Furthermore, overexpression of SETD7 attenuated the miR-345-5p induced anti-tumor effects on PTC cells.
Conclusions:
MiR-345-5p exhibited suppressive effects on PTC via targeting SETD7.
This study aimed to explore the effects of miR-345-5p on papillary thyroid carcinoma (PTC) and uncover the possible mechanisms.
Material and methods:
MiR-345-5p and SETD7 mRNA levels were analyzed by quantitative real-time PCR and SETD7 protein level was measured by Western blot. The viability, colony formation ability and apoptosis of PTC cells were measured with CCK-8, soft agar colony formation and flow cytometry assay, respectively. Luciferase reporter assay was used to identify miR-345-5p’s target.
Results:
Compared to neighboring normal tissues, there was lower miR-345-5p expression and higher SETD7 expression in PTC tissues. Moreover, Spearman’s correlation analysis indicated that there was a negative correlation between miR-345-5p and SETD7 expression in PTC tissues. MiR-345-5p mimics inhibited the viability and colony formation of TPC1 and B-CPAP cells and promoted apoptosis, whereas anti-miR-345-5p promoted PTC cell proliferation and inhibited apoptosis. SETD7 was confirmed to be a direct target of miR-345-5p through target scan analysis and luciferase reporter assay. Additionally, overexpression of SETD7 promoted the viability and colony formation of TPC1 and B-CPAP cells and inhibited apoptosis, whereas downregulation of SETD7 by shRNAs had opposite effects on PTC cells. Furthermore, overexpression of SETD7 attenuated the miR-345-5p induced anti-tumor effects on PTC cells.
Conclusions:
MiR-345-5p exhibited suppressive effects on PTC via targeting SETD7.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.