Introduction
Liver cancer is one of the most common malignant neoplasms around the globe. Its main classification is hepatocellular carcinoma (HCC), which poses a great threat to human health [1]. The main therapy strategy for HCC includes surgical operation, system radiotherapy, and chemotherapy [2]. Nevertheless, HCC cells have forceful proliferative activity and resistance to radiation and chemotherapy, which might be a reason for the inevitable recurrence and poor prognosis of HCC [3, 4]. Accordingly, it is urgent to study the molecular mechanisms involved in the tumorigenesis and chemoresistance of HCC, which will contribute to the progress of clinical therapy of HCC.
MicroRNAs are a class of small non-coding RNAs, which have been found to have various functions and complicated regulatory mechanisms, and they take part in almost all physiological or pathological cytobiological characteristics, including apoptosis, cell proliferation, and chemotherapy resistance [5–8]. Some studies have reported that MiRNA-610 was low-expressed in various malignant tumours, including HCC, colorectal cancer, and gastric cancer [9–11]. MiRNA-610 functions as a tumour suppressing gene, which can specifically silence many target genes to take part in tumour cell characteristics such as apoptosis and proliferation [12]. However, whether MiRNA-610 is involved in chemoresistance remains unclear.
We aim to study the role and mechanisms of MiRNA-610 in chemoresistance of HCC. This may offer a new therapeutic target against HCC.
Material and methods
Tissue specimens
All 76 HCC and control normal liver tissues (NLT) were obtained from Shengjing Hospital of China Medical University between Febraury 2010 and May 2011. The Ethics Committees of China Medical University approved this study, and handwritten permissions of patients were obtained. The clinical pathological data were confirmed by 2 pathologists according to guidelines for the standardised pathological diagnosis of primary liver cancer in China, and follow-up continued 5 years until April 2016.
All patients included 49 men and 27 women, whose ages ranged from 41 to 78 years. There were 37 patients with poorly differentiated and 39 patients with well or moderately differentiated tumour. Based on the seventh edition UICC/TNM classification, there were 44 patients in stage T1, 26 patients in T2, 6 patients in T3, 62 patients in N0, 14 patients in N1, 71 patients in M0, and 5 patients in M1.
Cell culture
Human liver HL7702 cell line and HCC HepG2 cell line were obtained from China Academy of Chinese Medical Sciences and cultured in DMEM medium with 10% foetal bovine serum (Gibco, USA) at 37° with 5% CO2.
Quantitative real-time PCR
Total RNA was extracted with Trizol reagent and cDNA synthesised with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Then MiRNA-610 and HDGF expression level was detected with SYBR (Applied Biosystems, Foster City, CA, USA). The primers of HDGF were 5’-GAGGGTGACGGTGATAAGAA-3’ (forward) and 5’-GAAACATTGGTGGCTACAGG-3’ (reverse). U6 and GAPDH were selected to be endogenous reference genes. The relative expression was quantified with the relative quantitative method [13].
Vector construction and transfection
The MiRNA-610 agonist (agomir-610) and negative control (agomir-NC) were purchased from Thermo Fisher Scientific (Foster City, CA, USA). The expression vector (pc-HDGF) and negative control vector (pc-NC) were constructed by Genscript (Nanjing, China). 50 nM agomir-610 or agomir-NC and 1.5 μg pc-HDGF or pc-NC were transfected with LipofectamineTM 3000 Reagent (Invitrogen, Foster City, CA, USA) according to the operating manual. After 4 h, normal media was applied to cultured cells with 48 h.
Cell proliferation assay
The cells (2 × 104/well) were seeded in a 96-well plate. The 96-well plate was given an additional 20 μl MTT solution (0.5 mg/ml, Sigma-Aldrich, USA) per well, and incubated at 37oC. Four hours later, the MTT solution was abandoned, and then to each well was added 0.2 ml followed by incubation for 30 min. The 96-well plate was read on an enzyme-labelled instrument (Bio-Rad Laboratories, Inc., Hercules, CA, USA) at 570 nm wavelength, and the calculation of cell viability refers to the previous literature [13].
Apoptosis detection
Flow cytometry was used to examine the apoptosis rate with an Annexin V-FITC apoptosis test kit (Beyotime, Nantong, China) according to the operating manual. Next, the cells were read by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA) (Ex, 488 nm; Em, 635 nm), and the data were analysed through CELLQuest 3.0 software (BD, USA). Cells in the right lower quadrant were regarded as apoptosis.
Chemotherapy resistance assay
Cells were treated with cisplatin (DDP) at various concentrations (0.1 μg/ml, 1 μg/ml, 5 μg/ml, 10 μg/ml, 25 μg/ml, 50 μg/ml) [14]. The cell viability was detected by MTT assay 48 h later and a dose-response curve was plotted. The half maximal inhibitory concentration (IC50) was calculated by probit regression model.
Luciferase reporter assay
The wild-type and mutant-type luciferase reporter vector (pmiR-HDGF-wt and pmiR-HDGF-mut) were constructed by the Genescript company (Nanjing, China) which included wild-type or mutant binding site of HDGF 3’UTR. The microRNA and luciferase reporter vector were cotransfected into HepG2 cells. The luciferase reporter assay was processed referring to the previous literature [13], the Dual-Glo luciferase assay (Promega Corporation, Madison, WI, USA) was applied to quantitate the relative luciferase activity, which was computed by the luciferase activity of Firefly to Renilla.
Western blot
Protein samples were harvested from cells, and were tested and treated with polyacrylamide gels electrophoresis, and then transferred to PVDF membrane. The membrane was hybridised first with Rabbit polyclonal HDGF antibody (ab176697, Abcam, USA) or Rabbit polyclonal GAPDH (ab9485, Abcam, USA) and then hybridised with secondary Goat Anti-Rabbit antibody (A0208, Beyotime, China) alternately. After that, the membrane was incubated with BeyoECL Plus reagent (Beyotime, Nanjing, China), and the images were obtained and analysed to quantify the expression of HDGF protein.
Statistical analysis
All experiments were repeated five times. The statistical analysis was completed by SPSS 23.0 software (IBM, USA). Differences were assessed by one-way analysis of variance (ANOVA) in nonparametric statistics and Student’s unpaired t-test. The correlation between the expression of MiRNA- 610 and HDGF was analysed by Spearman’s rank test. P < 0.05 means statistical difference.
Results
The low-expression and clinical significance of MiRNA-610
As shown in Figure 1 A, the MiRNA-610 expression in HCC specimens was much lower than that in NLT specimens (p < 0.05). Moreover, the MiRNA-610 expression in HepG2 cells was down-regulated compared with HL7702 cells (Figure 1 B, p < 0.05). Also, further statistical analysis indicated that low-expression of MiRNA-610 was more common in HCC patients with poor differentiation and high T stage (p < 0.05), but the expression of MiRNA-610 was not related with sex, age, positive lymph node metastasis, and distal metastasis (Table I). Those findings preliminarily proved that MiRNA-610 might be involved in the genesis and progression of HCC, but not in metastasis.
Table I
Correlation between miR-610 expression and clinical pathological parameters of hepatocellular carcinoma patients (n = 76)
| Parameter | Relative miR-610 expression | |||
|---|---|---|---|---|
| Number | Low | High | P-value | |
| Gender: | 0.2308 | |||
| Male | 49 | 27 | 22 | |
| Female | 27 | 11 | 16 | |
| Age: | 0.6445 | |||
| ≤ 50 | 42 | 22 | 20 | |
| > 50 | 34 | 16 | 18 | |
| Differentiation: | 0.0211* | |||
| Well, moderately | 39 | 16 | 26 | |
| Poorly | 37 | 22 | 12 | |
| T stage: | 0.0150* | |||
| T1 | 44 | 16 | 28 | |
| T2 | 26 | 17 | 9 | |
| T3 | 6 | 5 | 1 | |
| Lymph node metastasis: | 0.2366 | |||
| Positive | 14 | 9 | 5 | |
| Negative | 62 | 29 | 33 | |
| Distal metastasis: | 0.1651 | |||
| Positive | 5 | 4 | 1 | |
| Negative | 71 | 34 | 37 | |
Figure 1
MiRNA-610 was down-regulated in hepatocellular carcinoma (HCC). A, B – Relative expression of MiRNA-610 and hepatoma-derived growth factor (HDGF) in HCC tissues and cell lines. Compared to NLT or HL7702 cells, the MiRNA-610 expression was lower in HCC tissue and HepG2 cells, and HDGF showed significant downregulation. C – Kaplan-Meier analysis indicated that HCC patients with lower MiRNA-610 expression (n = 38) had a significantly worse prognosis than patients with higher MiRNA-610 expression (n = 38). *p < 0.05
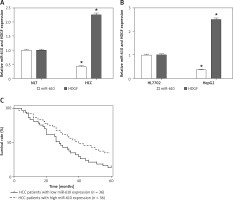
Figure 1 C shows that patients with higher MiRNA-610 expression had a remarkably longer survival time than patients with lower MiRNA-610 expression. All patients were divided into higher and lower MiRNA-610 expression groups from the median value of relative MiRNA-610 expression. The overall 5-year survival rate of is 37.5% in patients with higher MiRNA-610 expression but 18.5% in patients with lower MiRNA-610 expression. Meanwhile, patients with lower expression of MiRNA-610 had a significantly worse prognosis by univariate Cox proportional hazards regression analysis of overall survival (HR = 0.6867, 95% CI: 0.085–1.288; p = 0.036). The outcomes verified that MiRNA-610 was a prognostic factor for HCC patients.
MiRNA-610 played a tumour-suppressing role in HepG2 cells
In order to validate the effect of MiRNA-610 on proliferation ability and apoptosis of HepG2 cells, the agomir-610 was transfected into HepG2 cells to up-regulate the MiRNA-610 expression.
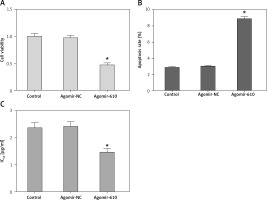
In comparison to the respective NC groups, the cell viability of HepG2 cells with MiRNA-610 over-expression decreased significantly (Figure 2 A, p < 0.05). The results of flow cytometry revealed that MiRNA-610 over-expression promoted the early apoptosis from 2.89% to 8.67% (Figure 2 B, p < 0.05), but the late apoptosis (from 10.37% to 11.46%) and necrosis (from 0.64% to 0.83%) showed no significant changes (p > 0.05).
Upregulation of MiRNA-610 inhibited DDP resistance in HepG2 cells
DDP is one of the first-line drugs in clinical chemotherapy for HCC patients. Figure 2 C shows how MiRNA-610 over-expression could depress IC50 of DDP from 2.37 ±0.19 to 1.47 ±0.13 μg/ml in HepG2 cells (p < 0.05), which demonstrates that over-expression of MiRNA-610 inhibits DDP resistance in HCC cells.
HDGF was a target gene of MiRNA-610 in HepG2 cells
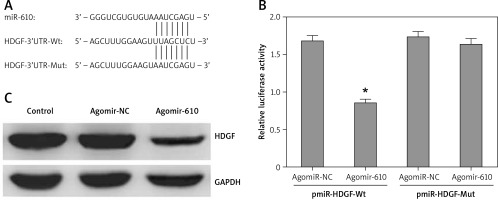
A conserved MiRNA-610 binding site was predicted in the 3’-UTRs of HDGF at 635-642 bp by several bioinformatics software packages, including TargetScan (Figure 3 A).
Figure 3
Hepatoma-derived growth factor (HDGF) was a target gene of MiRNA-610 in HepG2 cells. A – 3’-UTR region of HDGF mRNA is partially complementary to MiRNA-610. B – The effect of agomir-610 on the luciferase activity of pmiR-HDGF-3’UTR and pmiR-HDGF-3’UTR mutation. Firefly luciferase activity was normalised to Renilla luciferase. C – The influence of transfection with agomir-610 to HDGF protein expression in HepG2 cells. The relative quantitative analysis of HDGF protein normalised to GAPDH protein. *p < 0.05
The HDGF expression was up-regulated in HCC tissues compared with NLT tissues (Figure 1 B). Moreover, a negative correlation was identified by Spearman’s rank test between MiRNA-610 and HDGF.
Furthermore, luciferase reporter assay was used to investigate the above-mentioned prediction and the direct MiRNA610-HDGF interaction. Results showed that MiRNA-610 could inhibit the relative luciferase activity by combining with wild-type binding site, but not to mutant binding site (Figure 2 B, p < 0.05). However, the agomir-NC had no similar regulatory effects.
MiRNA-610 overexpression depressed HDGF protein expression in HepG2 cells as detected by western blot (Figure 3 C).
HDGF-mediated regulation induced by MiRNA-610 over-expression in HepG2 cells
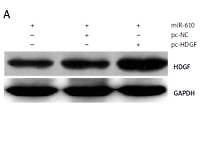
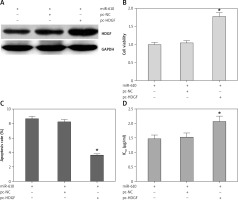
MiRNA-610 depressed the proliferation and DDP resistance of HepG2 cells and the HDGF gene was the MiRNA-610’s target, so we assumed HDGF-mediated regulation induced by MiRNA-610 over-expression in HepG2 cells. The transfection with pc-HDGF could up-regulate the expression of HDGF protein silenced by MiRNA-610 enhancement (Figure 4 A). HDGF over-expression in HepG2 cells reversed the MiRNA-610’s regulation on cell proliferation, apoptosis, and DDP resistance (Figure 4 B–D). In view of the above, we identified that MiRNA-610 depressed the proliferation and DDP resistance of HepG2 cells through targeted silencing of HDGF.
Figure 4
Hepatoma-derived growth factor (HDGF)-mediated MiRNA-610-induced regulation in HepG2 cells. A – The influence of transfection with agomir-610 and pc-HDGF on HDGF protein expression in HepG2 cells. B–D – HDGF overexpression reversed MiRNA-610’s regulatory roles in proliferation (B), apoptosis (C), and chemotherapy resistance (D) of hepatocellular carcinoma cells. *p < 0.05
Discussion
It is well known that microRNAs can regulate their target genes to participate in almost all cell biological characteristics, such as apoptosis and proliferation, which advances HCC development and results in poor prognosis [15–17]. Our study concluded that MiRNA-610 was down-regulated in HCC tissues and cell line, and lower MiRNA-610 expression in HCC tissues was associated with poor differentiation and higher T stage of HCC, but not with N and M stages. The results indicate that MiRNA-610 might also be involved in tumourigenesis and progress of HCC but has no role in metastasis. This is in concordance with a previous study in which MiRNA-610 down regulated in HCC and was involved in tumorigenesis through Wnt/β-catenin signalling pathway [9], which is the only report about MiRNA-610 in HCC.
Several MiRNAs, in addition to MiRNA-610, serve as prognostic factors for HCC patients, as with MiRNA26a/29a and MiRNA-454 [18, 19], which were associated with overall survival of HCC patients and could be prognostic factors for patients with HCC.
Experiments found that MiRNA-610 upregulation inhibited cell proliferation and advanced the cell apoptosis rate of HepG2 cells. Hence, our study demonstrated that MiRNA-610 serves as a tumour suppressor in cases of HCC.
Chemotherapy is an effective approach to prevent metastasis and recrudescence of HCC [20]. Chemoresistance is very common and is mainly responsible for failure of clinical treatment and poor prognosis. However, we report in this study that MiRNA-610 upregulation inhibits chemoresistance to DDP in HepG2 cells.
Given MiRNANAs specifically regulates their targeted genes to exert biological behaviours, such as MiRNA-147a and HIF-1α, MiRNA-181 and SAMHD1 [21, 22], we speculated that MiRNA-610 played its tumour-suppressor roles by silencing some target genes. The potential target genes for MiRNA-610 were predicted by several bioinformatic software packages [23], and the HDGF gene is the most intriguing one among these potential targets. HDGF was originally purified from the conditioned culture medium of human hepatoma-derived HuH-7 cell line [24]. HDGF was firstly identified for its oncogene roles in 2003 [25, 26]. More evidence certified the high expression of HDGF in many kinds of tumours and its relationship to tumour malignant progression and poor prognosis [27, 28].
A series of gain-of-function experiments affirmed that the HDGF gene was a target gene of MiRNA-610. First, the expression of HDGF was related negatively with the expression of MiRNA-610 in HCC tissues and cell line. Second, the specific combination between HDGF mRNA and MiRNA-610 had been proven effectively by luciferase reporter assay. Third, HDGF protein was silenced by MiRNA-610 enhancement.
Because MiRNA-610 overexpression suppressed the proliferation and DDP resistance in HCC cells, and HDGF is a MiRNA-610 target gene, it was suggested that MiRNA-610 plays its tumour-suppressive roles through silencing of HDGF. In order to testify the theory that transfection of pc-HDGF up-regulated the expression of HDGF silenced by MiRNA-610 enhancement in HCC cells, the following research discovered that HDGF over-expression reversed the regulatory roles of MiRNA-610 on proliferation, apoptosis, and DDP resistance of HCC cells. Accordingly, HDGF is an MiRNA-610 target gene and is involved in the regulatory process of MiRNA-610 in HCC cells.
In conclusion, MiRNA-610 not only plays a tumour suppressor role in HCC, but also affects chemoresistance to DDP. This role is mainly mediated through targeted silencing of the HDGF gene, which may offer a new potential therapeutic target and improve the clinical therapeutic effect for HCC.