Introduction
Abnormal levels of plasma lipids have been shown to be significantly associated with colon carcinoma risk [1–8]. Higher levels of low-density lipoprotein (LDL) cholesterol (LDL-C) have been reported to correlate with higher prevalence [9, 10] and risk [11, 12] of colon cancer. LDL-C is mainly cleared from the bloodstream by the liver’s LDL receptors (LDLRs). Regulation of LDLR expression has an important role in LDL-C hemostasis. Proprotein convertase subtilisin/kexin 9 (PCSK9) is a liver secretory protein that acts as a key regulator of protein levels of LDLRs, by which plasma levels of LDL-C in the body can be controlled. The secreted PCSK9 circulates in the bloodstream and binds to the cell surface LDLR. PCSK9 binding leads to internalization and degradation of LDLR in the lysosomal compartment and thereby reduces recycling of LDLR to the cell surface [13, 14]. Consequently, there are not enough LDLRs to remove plasma LDL-C when the circulating level of PCSK9 is increased as a result of gain-of-function mutations [15], whereas low levels of plasma PCSK9, due to loss-of function mutations, will lead to more intact LDLRs which in turn take up more plasma LDL-C [15]. Therefore, PCSK9 inhibition has emerged as a therapeutic tool and is currently used as an effective LDL-lowering approach in hypercholesterolemic patients [16–23]. Although the safety of PCSK9 inhibitors in hypercholesterolemic conditions has been approved, there is scant information on the safety of PCSK9 inhibition in other conditions such as cancer. To date, several human studies have been conducted to evaluate a possible link between PCSK9 gene variants and cancer risk, but the results have been inconsistent. One of the studies showed that there is a significant association between LDL-increasing PCSK9 mutations and a higher risk of cancer [24], while another human study could not show any association between PCSK9 loss-of-function mutations and incidence of cancer [25]. Conversely, a recent Mendelian randomization study showed that PCSK9 with LDL-increasing mutations is strongly correlated with higher cancer risk, whereas LDL-lowering variants mimicking PCSK9 inhibitors were found to be significantly associated with a lower risk of cancer occurrence [26]. These contradictory reports call for further investigations to assess the safety and efficacy of PCSK9 inhibitors in cancer. Anti-PCSK9 vaccines are a new generation of PCSK9 inhibitors, whose LDL-lowering effect is frequently verified in preclinical studies [27–30]. In a previous study, we reported a nanoliposomal anti-PCSK9 vaccine with long-lasting, specific and safe inhibitory effects on plasma PCSK9 in BALB/c mice [31].
In the current study, we aimed to investigate the effects of PCSK9 inhibition using the a nanoliposomal anti-PCSK9 vaccine in BALB/c mice bearing CT26 colon carcinoma.
Material and methods
Vaccine preparation and characterization
Preparation and characterization of the liposome nanoparticles
The thin film-lipid hydration method was used to synthesize a nanoliposome formulation containing 1,2-dimyristoyl-sn-glycero-3-phosphorylcholine (DMPC), 1,2-dimyristoyl-sn-glycero-3-phosphoryl-glycerol (DMPG), and cholesterol (Chol) (Avanti Polar Lipid; Alabaster, USA) at a final concentration of 40 mM (total phospholipids and Chol). In brief, DMPC, DMPG, and Chol were mixed in chloroform at the molar ratios of 75 : 10 : 15, respectively. Lipid solution was dried to a thin lipid film under reduced pressure using rotary evaporation (Heidolph, Germany), and then the organic solvent was completely eliminated using over-night freeze drying (VD-800F, Taitech, Japan). Afterward, the dried lipids were dispersed using hydration with 10 mM HEPES buffer (pH 7.2) containing 5% dextrose, followed by vortexing and bath-sonicating to be completely homogenized. The obtained multilamellar vesicles (MLVs) were serially extruded using a mini extruder (Avestin, Canada) with polycarbonate membranes of 600, 400, 200, and 100 nm pore size, respectively, to prepare small unilamellar vesicles (SUVs) with a uniform size of 100 nm. Particle size (diameter, nm), zeta potential (surface charge, mV) and polydispersity index (PDI) of the prepared nanoliposomal formulation were evaluated using dynamic light scattering (DLS) technique on a Zetasizer (Nano-ZS, Malvern, UK) at room temperature (RT). The prepared liposome nanoparticles were stored at 4°C under argon.
Preparation of immunogenic peptide
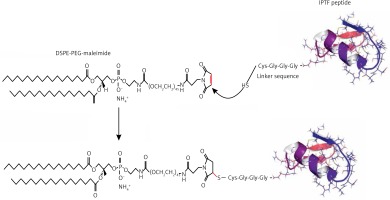
The Immunogenic Fused PCSK9-Tetanus (IFPT) peptide with a purity grade of > 95% was synthesized and high performance liquid chromatography (HPLC)-purified by ChinaPeptides Co., Ltd. (Shanghai, China). The already designed IFPT construct [31] contains a PCSK9 peptide, as a B cell epitope inspired from the AFFiRiS group [27, 32], and a T-helper cell epitope belonging to tetanus toxin included as a standard adjuvant carrier [33] (Table I). To link the IFPT epitope on the surface of liposome nanoparticles, it was bound to DSPE-PEG-Mal (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(PEG)-2000]) lipid (Lipoid GmbH, Germany) via an N-terminal cysteine residue inserted in IFPT peptide (Figure 1).
Table I
Sequence of immunogenic peptides used in the present study
| Peptide name | Sequence | Immunogenicity |
|---|---|---|
| PCSK9 | S-I-P-W-N-L-E-R-I-T-P-V-R | B cell epitope |
| Tetanus | A-Q-Y-I-K-A-N-S-K-F-I-G-I-T-E-L | T cell epitope |
| IFPT | *CGGGSIPWNLERITPVRKKAQYIKANSKFIGITEL |
Manufacturing of DSPE-PEG-IFPT micelles
DSPE-PEG-Maleimide lipid was used to link the IFPT peptides on the surface of liposome nanoparticles as an adjuvant delivery system. The N-terminal cysteine residue of the IFPT peptide provides a thiol group that reacts with the pyrrole group maleimide and generates a thioether bond, whereby the peptide covalently conjugates with the DSPE-PEG-Maleimide linker. The IFPT peptide and DSPE-PEG-Mal at the molar ratios of 1.2 : 1, respectively, were dissolved in DMSO/chloroform solution at the volume ratio of 1 : 1, and then gently stirred at RT for 48 h. The linkage was confirmed using the TLC (thin layer chromatography) method with the mobile phase containing chloroform, methanol, and water at the volume ratio of 90 : 18 : 2. After that, the DMSO/chloroform solution was dried by a rotary evaporator and freeze-drying followed by hydration with sterile deionized water (pH 7.2) at 30°C to prepare DSPE-PEG-IFPT micelles. The efficiency of the linkage in the constructed micelles was estimated by HPLC analysis. The true value of the linked micelles was measured using efficiency of linkage and content of total lipid determined by the Bartlett phosphateassay method [34].
HPLC analysis of linkage efficiency
The efficinecy of linkage between the IFPT peptide and the DSPE-PEG-Mal linker was indirectly estimated by HPLC quantification of the free peptide content of the prepared DSPE-PEG-IFPT micelles. The HPLC apparatus was equipped with a Smart line HPLC Pump 1000, a PDA Detector 2800 (set at 220 nm), and a Degasser5000, all from Knauer (Berlin, Germany). Each sample (20 μl) was injected through a Smart line auto sampler and data were obtained and processed with ChromGate software (version 3.3.1) from Knauer (Berlin, Germany). Chromatographic separation was performed on a C18 reverse-phase column, 4.6 mm × 25 cm (Shimadzu, Japan), using an isocratic mobile phase of (0.1% TFA in water)/(0.1% TFA in acetonitrile) at gradient ratios of 55/45 to 45/55 in 10 min, at a flow rate of 1 ml/min.
The IFPT peptide with HPLC purity > 95% was used as a standard solution. The free peptide peak within the chromatogram of the micelle sample was identified and assigned based on the retention time (2.2 min) of the standard solution, followed by sample spiking.
To quantify the free peptide content of the micelle sample, a calibration curve was constructed by injecting standard solution at five concentrations (50–500 μg/ml), which was linear with a correlation coefficient (r 2) of 0.9954. Using linear regression analysis of the calibration curve appearing in the standard chromatogram, the free peptide content of the micelle sample was measured. Linkage efficiency in the constructed DSPE-PEG-IFPT micelles was calculated by subtracting the free peptide amount within the micelles quantified by HPLC from the amount of the IFPT peptide initially added.
Construction and cahraacterization of nanoliposomal IFPT vaccine
Nanoliposomes were used as a delivery adjuvant to enhance immunogenicity of the peptide. Since many IFPT peptides can be conjugated to the surface of liposome nanoparticles, we propose that IFPT-linked nanoliposomes can elicit a high-titer antibody against self-antigen PCSK9, maybe through elevating peptide valency. To attach the IFPT peptide on the nanoliposome surface, the post-insertion approach was performed, in which the prepared DSPE-PEG-IFPT micelles (100 μg, based on the linked peptide) and liposome nanoparticles (1 ml) were mixed and then gentely shaken at 45°C for 3 h. The micelles were inserted in the nanoliposome bilayer via the DSPE phospholipid moiety, and exposed IFPT peptides on the nanoliposome surface through the PEG chains. Particle size, surface charge and PDI of the prepared nanoliposomal IFPT particles were evaluated using DLS technique on a Zetasizer (Nano-ZS, Malvern, UK) at RT. The IFPT-conjugated nanoliposomes were adsorbed to 0.4% Alum adjuvant (Sigma-Aldrich) at a 1 : 1 (v : v) ratio in a total volume of 400 μl and stored at 4°C under argon. Prior to injection, the nanoliposomal IFPT plus Alum vaccine, hereafter called L-IFPTA+, was brought to RT and carefully mixed.
Animal and cell line
A total of 15 female BALB/c mice (4–6 weeks old) were purchased from Pasteur Institute of Tehran, Iran and fed with ad libitum access to purified water and a commercial stock diet. All mice were housed in a pathogen-free animal house at a temperature of 22 ±1°C with a 12 : 12 h light : dark cycle and maintained under a relative humidity of 50 ±10%. Animal care was performed in accordance with welfare guidelines established by the Institutional Ethical Committee and Research Advisory Committee of Mashhad University of Medical Sciences. At the end of the experiment all animals were euthanized by CO2 inhalation.
The CT26 colon carcinoma cell line was provided from Pasteur Institute of Tehran, Iran and cultured in RPMI-1640 medium containing 10% FBS and supplemented with penicillin (100 IU/ml)/streptomycin (100 mg/ml). The cells were incubated at 37°C with a 5% CO2/95% air humidified atmosphere.
Vaccination schedule
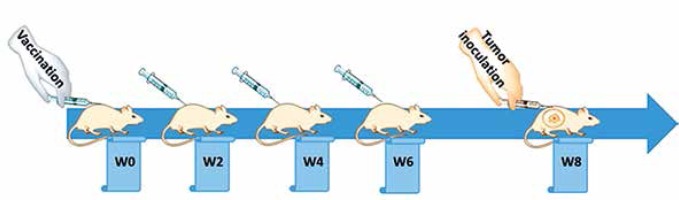
Following one week of taming prior to the experimental procedures, the mice were randomly arranged into two groups: a vaccine group (n = 5) and an untreated group (n = 10) group. The vaccination was subcutaneously primed at week 0 (W0) and followed by three boosters (W2, W4, and W6), in bi-weekly intervals (Figure 2), while untreated mice simultaneously received phosphate-buffered saline (PBS). Tail vein bleeding was performed two weeks after each vaccination for the titration of plasma anti-PCSK9 antibody.
Evaluation of plasma anti-PCSK9 antibody
To measure the titer of anti-PCSK9 antibodies, plasma samples were collected and assayed by the ELISA method. Briefly, PCSK9 peptide at the concentration of 5 μg/ml in 0.1 M NaHCO3 (pH 9.2–9.4) was coated overnight in a 96-well Nunc-MaxiSorp plate. Free binding sites were then blocked by incubation with blocking buffer (1× PBS, 1% BSA) for 1 h at 37°C. Diluted plasma (1 : 400 in 1× PBS/0.1% BSA/0.05% Tween-20) was added, serially diluted 1 : 4, and incubated for 1 h at 37°C. Each ELISA plate contained a standard antibody as an internal control. For detection, biotinylated anti-mouse IgG (H + L) (Sigma-Aldrich; 1 : 1000) in 1× PBS/0.1 % BSA/0.1% Tween-20 was applied and incubated for 1 h at 37°C. Then, horseradish peroxidase coupled to streptavidin (Roche) was added (30 min, 37°C) followed by the addition of the substrate 2,2′-azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt (ABTS) (Sigma-Aldrich) (15 min, RT). The optical density (OD) at 450 nm was measured with a Microwell plate reader (Sunrise, Tecan, Switzerland) and the titers were defined as the dilution factor referring to 50% of the maximal optical density (ODmax/2). The mean titers ± SD of all animals per group are presented.
Plasma PCSK9 quantification
Plasma PCSK9 concentration in the vaccinated mice was measured by CircuLex rat PCSK9 ELISA (CircuLex, Cy-8078, MBL, Woburn, MA) according to the manufacturer’s instructions. Briefly, 100 μl of the diluted 1 : 100 plasma samples was added to a 96-well microplate and incubated for 1 h at RT. A HRP-conjugated anti-PCSK9 antibody was added for 1 h followed by the substrate reagent and stop solution, all at RT. Optical density was detected at 450 nm with a Microwell plate reader (Sunrise, Tecan, Switzerland). A standard curve provided by the supplier was defined to measure PCSK9 concentration.
PCSK9 inhibition analysis
To assay inhibition of mice PCSK9 by vaccine-induced antibodies, interaction of induced antibodies with PCSK9 was evaluated. For this purpose, the same kit CircuLex rat PCSK9 ELISA was used, but for HRP-conjugated anti-PCSK9 antibody, detection was performed with HRP-conjugated anti-mouse IgG (H + L) (Sigma Aldrich; dilution 1 : 5000) incubated for 1 h at RT, followed by the substrate reagent and stop solution provided by the supplier. The OD was detected at 450 nm with the Microwell plate reader.
In vitro evaluation of PCSK9-LDLR binding
CircuLex PCSK9-LDLR in vitro binding assay kit (CircuLex, Cy-8150, MBL, Woburn, MA) was used to evaluate the ability of vaccine-induced antibodies to inhibit the PCSK9-LDLR interaction in vitro. Briefly, 100 μl of vehicle control or the plasma samples of vaccinated mice was added to a 96-well microplate pre-coated with a recombinant LDLR-AB domain, which contains a binding site for PCSK9. Immediately after that, the reaction was initiated by adding a “His-tagged PCSK9 wiled type” solution incubated for 2 h followed by adding a biotinylated anti-His-tag monoclonal antibody for 1 h at RT. Then, HRP-conjugated streptavidin was coated for 1 h at RT followed by the substrate reagent and stop solution. In this method, the higher amount of PCSK9-LDLR interaction is associated with higher ELISA OD, in which in the presence of anti-PCSK9 antibody this interaction is inhibited and consequently ELISA OD is decreased. A dose-response curve with appropriate serial dilutions of “His-tagged PCSK9 wild type” solution was drawn to measure the accurate inhibition percentage of test samples.
Evaluation of in vivo anti-tumor efficacy
Two weeks after the last booster, the vaccinated and unvaccinated BALB/c mice were subcutaneously inoculated with CT26 colon carcinoma cells (5 × 105/50 μl PBS/mouse) into the right flank at day zero. Tumor growth was monitored in 3-day intervals by calculating the tumor volume after measuring three orthogonal diameters with calipers according to the formula: tumor volume (mm3) = (length × height × width) × 0.52. After the tumor mass became palpable (approximately 10 mm3) at day 10, the mice were randomly divided into three groups (5 mice/group) and subjected to different treatment protocols: (1) the vaccine group comprising vaccinated tumor-bearing mice that received a single tail vein injection of PBS, (2) the Doxil (liposomal doxorubicin) (positive control) group comprising unvaccinated tumor-bearing mice that received a single tail vein injection of Doxil (15 mg/kg), and (3) the control (PBS) group comprising unvaccinated tumor-bearing mice that received a single tail vein injection of PBS.
To evaluate therapeutic efficacy, mouse body weight, tumor size, general health, and survival were monitored every other day for 50 days. Animals’ euthanasia (CO2 inhalation) was performed on those with 4T1 tumor due to the following ethical considerations: body weight loss > 20% of initial mass, tumor volume greater than 2.0 cm in one dimension, or mice became sick and unable ambulate to reach food/water [35, 36]. For each mouse, the time to reach tumor volume above 1000 mm3 or the time to reach the end point (TTE) as a response variable was calculated from the equation of the line obtained by exponential regression of the tumor growth curve. For each group, the percent of tumor growth delay (%TGD) was determined by calculating the difference between the average TTE of the treatment group (T) and the average TTE of the control group (C), (%TGD = [(T – C)/C] × 100) [37]. For each treatment group, the percent of increased life span (%ILS) was measured based on the following formula: [(average survival time of treatment group / average survival time of control group × 100) – 100] [38].
Statistical analysis
Statistical analysis was performed using SPSS Statistics version 20 and GraphPad Prism version 7.04 software. Survival data were analyzed using the log-rank (Mantel-Cox) test. Other comparisons were done using one-way ANOVA and the Tukey post-hoc multiple comparison test. Values were expressed as mean ± SD or median for normally and non-normally distributed data, respectively. Results with p < 0.05 were considered as statistically significant.
Results
Physical properties
Characterization of liposome nanoparticles showed a size of 145 nm and 180 nm for free and IFPT-linked nanoliposomes, respectively. Other characteristics including PDI and zeta potential are detailed in Table II.
Analysis of DSPE-PEG-IFPT micelles
Attachment of the IFPT peptides to the DSPE-PEG-Mal linker was confirmed qualitatively and quantitatively using TLC (Figure 3) and HPLC (Figure 4) methods, yielding an attachment yield of 96% according to HPLC.
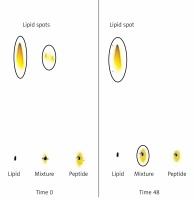
Figure 3
Assessment of conjugation between DSPE-PEG-Mal and the IFPT peptide at time zero and 48 h after start of reaction. Lipid (DSPE-PEG-Mal) is dissolved in the mobile phase and ascends to the top of the TLC plate (spots in the top of the left and middle lines) but peptide is bound to the silica and remains in the spotting point (the middle line). After 48 h, lipid bound to peptide and stayed at the point of spotting, and therefore the lipid spot on the top of the reaction mixture line disappeared, indicating the conjugation of the IFPT peptide and DSPE-PEG-Mal linker
L-IFPTA+ vaccine-induced PCSK9 antibody response in BALB/c mice
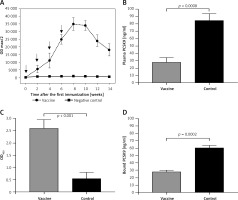
The L-IFPTA+ vaccine induced a strong IgG response against PCSK9 peptide in BALB/c mice upon 4 immunizations in biweekly intervals. Notably, long-term analysis revealed that anti-PCSK9 antibody titer peaked at week 8 and remained constant up to week 10, and then revealed a decreasing trend (Figure 5 A).
Figure 5
AntiPCSK9 vaccine efficacy. A – L-IFPTA+ vaccine could induce anti-PCSK9 antibody titers (ODmax/2) upon 4 immunizations in a bi-weekly interval (indicated by arrows). Antibody titers were assayed over 14 weeks after prime immunization. B – Concentrations of plasma PCSK9 in the vaccine and control group were 28 ±6 ng/ml and 85 ±9 ng/ml, respectively. C – Direct detection of antibodies bound to plasma PCSK9 in the plasma samples from vaccinated and control mice. Increased OD450 is indicative for vaccine-induced anti-PCSK9 antibodies which directly target PCSK9. D – In vitro PCSK9/LDLR binding assay. Plasma sample of vaccine group could decrease PCSK9 binding to LDLR by 53%, when compared with plasma sample of control group. Values are expressed as means ± SD (n = 3 replicates of the pooled samples of 5 mice). Significance compared to control values was analyzed by unpaired 2-tailed Student’s t-test
Targeting of plasma PCSK9 by vaccine-induced antibodies
Vaccine-induced PCSK9 antibodies targeted plasma PCSK9 in vaccinated mice in a specific manner. Plasma PCSK9 levels were 28 ±6 ng/ml and 85 ±9 ng/ml in the vaccinated and control groups, respectively (Figure 5 B). Plasma PCSK9 concentrations were decreased by 67% (–57 ±6 ng/ml, p = 0.0008) in the vaccinated mice as compared to control mice. The specificity of PCSK9 targeting by vaccine-induced antibodies was tested using an ELISA assay as described above and in our previous report [31]. A significantly higher OD450 signal was detected from the plasma of vaccinated mice, suggesting that vaccine-induced PCSK9 antibodies were able to directly bind to PCSK9 (Figure 5 C).
PCSK9-LDLR interaction blockade by vaccine-induced PCSK9 antibodies
Using the above-mentioned in vitro assay, PCSK9 antibodies were found to inhibit the interaction between PCSK9 and LDLR. Binding of PCSK9 to LDLR was reduced by 53% in the presence of plasma obtained from L-IFPTA+-vaccinated mice, as compared to control mice (Figure 5 D).
Efficacy of liposomal-antiPCSK9 in CT24 colon carcinoma model treatment
To assay the effect of PSCK9 inhibition on breast cancer progression, a CT26 colon carcinoma model was developed in BALB/c mice previously immunized with liposomal antiPCSK9 vaccine. The protective effect of the vaccine on tumor-bearing mice was evaluated by monitoring body weight alterations, tumor growth rate in terms of mean tumor size (mm3), and survival.
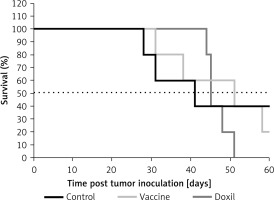
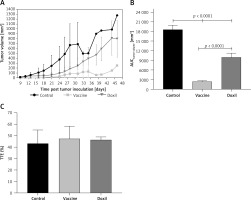
The weight monitoring curve showed that body mass was significantly reduced in the vaccinated mice after 25 days of tumor induction. As compared with the control group, vaccinated mice showed a significantly higher body weight loss from 25 days after tumor induction (p < 0.05) (Figure 6 A). Analysis of the integrated areas under the body weight curve (AUCbody weight) over 50 days showed that the body weight of vaccinated mice was decreased by 7.7% in comparison with the control mice, while there was no significant difference between vaccine and Doxil groups (Figure 6 B). To determine endpoint therapeutic modalities of antiPCSK9 vaccine in tumor-bearing BALB/c mice, time to reach endpoint (TTE) and percentage of tumor growth delay (%TGD), as well as median survival time (MST) and increase in life span (ILS), were measured (Table III). Monitoring the tumor size (Figure 7 A) revealed that three out of five vaccinated mice remained tumor-free, and tumor growth in the remaining two vaccinated mice was significantly lower than that in Doxil and the control mice. Data of the integrated areas under the tumor volume curve (AUCtumor volume) over 50 days demonstrated that tumor volume was 77% and 87.7% lower (p < 0.0001) in the vaccinated mice than in the Doxil and the control mice, respectively (Figure 7 B). TTE of the vaccine group (47 ±11 days) was slightly but not significantly higher than that of the Doxil (46 ±2.6 days) and control (43 ±12 days) groups. TGD as a widely accepted method for qualifying in-situ assessment of tumor treatment quality showed that the tumor growth rate in the vaccine and Doxil group was reduced by 9.3% and 7.3%, respectively, compared with the control group. Kaplan-Meier curves (Figure 8) showed that the vaccinated mice survived slightly but not significantly longer than the Doxil and the control mice (p < 0.05, log-rank test). The MST of the vaccine, Doxil and control group was 51, 45, and 41 days, respectively. Analysis of the ILS revealed that the vaccinated mice’s life was prolonged by 24.4% as compared with the control mice, while it was increased by 9.8% in the Doxil group (Table III).
Table III
Therapeutic efficacy data of different treatments in BALB/c mice bearing CT26 colon carcinoma
| Groups | TTE [days] Mean ± SD | TGD (%) | MST [days] | ILS (%) |
|---|---|---|---|---|
| Control | 43 ±12 | – | 41 | – |
| Vaccine | 47 ±11 | 9.3 | 51 | 24.4 |
| Doxil | 46 ±3 | 7.3 | 45 | 9.8 |
Figure 6
The weight monitoring curve (A) shows point-by-point changes of the body weight during 50 days in the control, vaccine, and Doxil group. The integrated areas under the body weight curve (AUCbody weight) over 50 days (B) demonstrates overall weight changes. Data with p < 0.05 were considered as statistically significant. The body weight loss was significantly (p < 0.0001) higher in the vaccine and Doxil groups than in the control group. Animal body weight was measured every 3 days
Data are presented as the mean ± SD (n = 5).
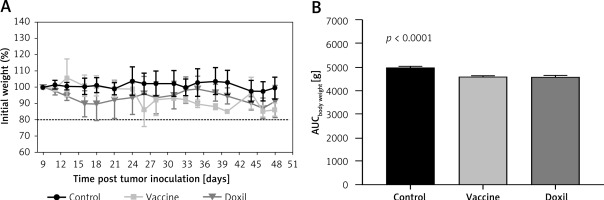
Figure 7
The tumor growth curve (A) shows the increase of tumor size during 50 days in the control, vaccine, and Doxil groups. Tumor volume (mm3) was measured every 3 days. The integrated areas under the tumor volume curve (AUCtumor volume) over 50 days (B) demonstrates overall increase of tumor size. Measuring the time to reach endpoint (TTE) or the time to reach tumor volume above 1000 mm3 (C) showed no significant difference between studied groups. Data with p < 0.05 were considered as statistically significant
Data are presented as the mean ± SD (n = 5).
Discussion
Recent Mendelian randomization studies analyzing the correlation of PCSK9 polymorphisms with cancer occurrence have shown contradictory results with respect to the association of LDL-rising/lowering PCSK9 mutations and risk of cancer [24–26]. However, there have been no data on the effect of PCSK9 inhibition on cancer progression. Dyslipidemia, particularly an increased plasma level of LDL-C, has been reported to be significantly correlated with a higher risk and incidence of colon cancer [9–12]. However, the effects of pharmacological LDL-C lowering on cancer has remained unclear. In this study, we explored whether PCSK9 inhibition has any positive, negative or neutral effect on the endpoints of colorectal cancer. Interestingly, our results showed that the nanoliposomal anti-PCSK9 (L-IFPTA+) vaccine, as a PCSK9 inhibitor, could moderately inhibit tumor development in mice bearing CT26 colon carcinoma.
L-IFPTA+ vaccine induced generation of functional anti-PCSK9 antibodies (Figure 5 A) that reduce plasma level and activity of PCSK9 (Figures 5 B, D) through specific and direct targeting of PCSK9 (Figure 5 C) in BALB/c mice. The vaccine-induced anti-PCSK9 antibodies were found to significantly reduce tumor growth (Figure 7), partially increase MST (Figure 8), and prolong lifespan (Table III) in the vaccinated mice bearing colorectal cancer, when compared with Doxil and control groups.
The moderate anti-tumor effect of the tested anti-PCSK9 vaccine is underpinned by the recent Mendelian randomization study which revealed that LDL-lowering mutations of PCSK9 mimicking PCSK9 inhibitors were correlated with a lower risk of breast cancer occurrence, while PCSK9 with LDL-raising mutations has a causal correlation with a higher risk of breast cancer [26]. It is further supported by another human study analyzing a phenome-wide association of LDL-lowering genetic variants in the PCSK9 locus mimicking PCSK9 inhibitors, showing that these PCSK9 variants do not increase the risk of cancer [39]. However, a prospective, population-based cohort study showed that there is no association between LDL-lowering PCSK9 polymorphisms and the risk of cancer [25].
In conclusion, the present data demonstrated that PCSK9 inhibition is potentially safe, and might moderately inhibit tumor progression in an experimental model of colon cancer. These findings can support the potential safety of PCSK9 inhibition in cancer, though further preclinical and clinical trials taking into account different PCSK9 inhibition approaches, longer term follow-up durations and other cancer models are necessary to confirm the safety and possible efficacy of PCSK9 inhibition in malignant conditions.