Introduction
Bladder cancer (BC) is a common urinary tract cancer and according to the global cancer statistics in 2012 there were 429,800 new BC cases and 165,100 BC related deaths [1]. Bladder cancer can be classified by its stage and pathological characteristics into 2 types, non-muscle invasive BC (NMIBC) and muscle invasive BC (MIBC). Approximately 70% of BC patients are diagnosed with superficial NMIBC, which can usually be treated through transurethral resection and has a high 5-year survival rate close to 90% [2]. Radical cystectomy is considered to be the primary treatment for MIBC despite the fact that it can lower patients’ quality of life. Under optimal conditions, organ-sparing chemoradiotherapy can be used as an alternative to radical cystectomy; however, even with this treatment the 5-year survival rate of MIBC patients is still only 60% [3]. The main cause of this low survival rate is distant recurrence [4, 5]. In the last 20 years, little progress has been made in the development of new treatment strategies for BC, particularly in MIBC [6]. Novel therapeutic methods are urgently required. The discovery of microRNA (miRNA) in the human genome has brought new insight into cancer research [7, 8].
MiRNAs are a class of small non-coding RNAs which have a close association with cell development, proliferation and metastasis in human cancers [9]. By binding to the 3′-untranslated region (3′-UTR) of the corresponding mRNA, miRNAs can cause mRNA degradation or translation suppression. MiRNAs can regulate gene expression and thus modulate various important cellular processes such as cell growth, proliferation, migration and apoptosis [10]. Depending on the gene they target, miRNAs can serve as either tumor suppressors or oncogenes in a variety of cancers [11, 12]. For example, miR-31 inhibits the proliferation and invasion of BC cells by regulating the expression of RAP2B, and miR-130 enhances cell migration by targeting PTEN in BC cell lines and tissues [13, 14]. Previous studies have demonstrated that miR-124 was down-regulated in various cancers including breast cancer, bladder cancer, retinoblastoma, glioblastoma and esophageal cancer [15–19]. Furthermore, several studies have pointed out the potential tumor suppressor role of miR-124 in bladder cancer [20, 21]. However, the functional roles and special molecular mechanisms behind miR-124-3p in bladder cancer are not completely understood, and thus further research is required.
Endothelin receptor type B (EDNRB) is a kind of G-protein-coupled receptor for endothelin-1 [22]. The combination of endothelin-1 and EDNRB is referred to as the endothelin-axis and has been associated with cell proliferation, migration and apoptosis [23, 24]. Furthermore, previous research has demonstrated that EDNRB was up-regulated in bladder cancer and the expression of EDNRB was associated with the prognosis of cancer [22]. Although EDNRB is a potential target gene of miR-124-3p, the association between miR-124-3p and EDNRB has not yet been reported. We aimed to reveal the relationship between miR-124-3p and EDNRB in bladder cancer.
In this study we aimed to uncover the mechanisms whereby miR-124-3p was down-regulated in BC cell lines and suppressed BC cell proliferation and invasion through regulation of EDNRB in detail. These results will support the theorized role that miR-124-3p has in bladder cancer and will provide an important clue in explaining the tumorigenesis of bladder cancer.
Material and methods
Tissue samples
Tumor and adjacent tissues were obtained from 27 BC patients at the First Affiliated Hospital of Guangxi Medical University between May 2015 and March 2016. Benign tumor tissues were obtained from 20 age- and sex-matched benign prostatic hyperplasia patients (pathological information of patients is shown in Table I). Tissue samples were collected during the radical cystectomy procedures and were immediately refrigerated in liquid nitrogen until their use in the experiment. The use of tissue samples was authorized by the ethics committee of the First Affiliated Hospital of Guangxi Medical University.
Table I
Pathological information of patients
Cell culture and transfection
Immortalized human bladder urothelium cell line SV-HUC-1 and human bladder cancer cell lines TCCSUP, J82 and 5637 (purchased from ATCC, VA, USA) were cultured in RPMI 1640 medium with 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin under standard culture conditions (37°C and 5% CO2). PCDH plasmids, miR-124-3p mimics (5′-UAAGGCACGCGGUGAAUGCC-3′-), miR-124 inhibitors (5′-CGUGUUCACAGCGGACCUUGAU-3′-), EDNRB siRNA (5′-CCATGGGAGTCTTTAGATG-3′-) and EDNRB cDNA were purchased from Shanghai GenePharma Co., Ltd.
TCCSUP cells were transfected when coverage reached 30-50% and then cells were divided into 5 groups. Cells were transfected using the Lipofectamine 2000 reagent (Invitrogen Corporation, USA). Cells in the control group did not undergo any transfection, cells in the NC group were transfected with a nonsense sequence, cells in the miR-124-3p mimics group were transfected with miR-124-3p mimics, cells in the miR-124-3p inhibitor group were transfected with miR-124-3p inhibitors, cells in the EDNRB siRNA group were transfected with EDNRB siRNAs, and cells in the miR mimics + EDNRB group were transfected with miR-124-3p mimics and EDNRB cDNA.
MTS assay
48 h after transfection, cells from the various groups were seeded into a 96-well plate (5 × 103 cells per well). After 24 h, 48 h and 72 h of cultivation, the cell proliferation was detected using Cell Titer 96 AQueous One Solution Cell Proliferation Assay (MTS) (Promega, Beijing, China). The absorption rate of the cells at 490 nm was measured using an ELISA reader.
Colony formation assay
After 48 h of transfection, cells were digested with pancreatic enzymes and forced into single-cell suspension. Cells were then seeded into six-well plates (500 cells per well) and cultured for 2 weeks. After the colonies were fixed and stained with crystal violet, colony counts were conducted.
Flow cytometry
For the cell cycle assay, 3 × 105 cells were collected from each group and fixed with 70% cold ethanol overnight at 4°C. Cells were then washed 3 times with PBS and treated with 100 μg/ml of RNase A for 30 min. Subsequently, cells were treated with 50 μg/ml of propidium iodide (PI) for 30 min in the dark and then the DNA distribution was measured using a FACScan flow cytometer (BD Biosciences, USA). The apoptosis rate of samples was calculated after the cells were stained using the Annexin V-FITC/PI Apoptosis Detection Kit (BD Biosciences) in flow cytometry.
Tumor formation in nude mice
Six-week-old BALB/c-A nude mice (purchased from the Laboratory Animal Center Southern Medical University) were obtained to build in-vivo tumor growth models. Mice were randomly divided into 4 groups with 5 mice in each group. Mice were then subcutaneously injected with 1 × 107 TCCSUP cells (1 × 107 cells per mouse). Seven days later when the tumor volumes were quantified, tumor sites were transfected with negative control, miR-124-3p mimics and EDNRB siRNA for 2 weeks via direct injection (2 × 107 units each time, twice a week). Tumor size was measured every 4 days according to the following formula: volume = (A × B 2)/2, where A and B were the largest and the smallest diameters, respectively. All mice were sacrificed at the end of the observation period (24 days after cell injection).
Dual-luciferase reporter assay
The wild type EDNRB 3′-UTR sequence was amplified using RT-PCR and inserted into the pGLO luciferase carrier (Promega Corporation, USA), as pGLO-EDNRB-3′-UTR wt. In the mutant type plasmid, pGLO-EDNRB-3′-UTR mut, the complementary sequences for miR-124-3p at EDNRB 3′-UTR were mutated using the site mutation method. The luciferase carrier was directly transfected into TCCSUP cells in combination with either miR-124-3p mimics or the negative control respectively plus Lipofectamine 2000 (Invitrogen, USA). 48 h after transfection, the DLR dual luciferase reporter assay system (Promega Corporation, USA) was used to measure the luciferase activity in cells.
Real-time quantitative PCR (qRT- PCR)
Total RNA from tissue and cell samples was extracted according to the instructions using the Trizol reagent (Invitrogen, USA). CDNA was then synthesized using the mRNA and miRNA reverse transcription kits (Takara Biotechnology Co., Ltd). Finally, cDNA was used as the template to measure qRT-PCR reactions with the PCR kit (Takara Biotechnology Co., Ltd). U6 and GAPDH served as the internal controls for miRNA and mRNA respectively. The relative expression levels of miRNA and mRNA were recorded using 2–ΔΔCt values. Primer sequences are shown in Table II.
Western blotting analysis
Total protein was dissociated from tissue and cell samples and then protein density was detected using the BCA protein assay. After protein electrophoresis, membranes were blocked with 5% skimmed milk at room temperature for 4 h, incubated for 1 h at 37°C with anti-EDNRB, CBL, STAT3 primary antibodies which had been diluted with confining liquid and then washed 4 times with tris-buffered saline Tween (TBST). Subsequently, membranes were incubated at 37°C for 1 h with secondary antibody (anti-Rabbit IgG) coated with horseradish peroxidase (HRP) and rewashed 4 times with TBST. With reduced glyceraldehyde-phosphate dehydrogenase (GAPDH) as the endogenous control, the samples were ultimately handled with enhanced chemiluminescence (ECL) for 5 min and images were captured after exposure in a darkroom.
Statistical analysis
All data analysis was performed using the SPSS 18.0 statistical software (Chicago, Illinois, USA) and the results are presented as mean ± standard deviation (mean ± SD). Normally distributed measurement data were compared using the t test or analysis of variance (ANOVA). Measurement data which were not normally distributed were compared using the nonparametric rank-sum test. A p-value less than 0.05 was considered to be statistically significant.
Results
Expression of miR-124-3p and EDNRB in cancer tissues and cells
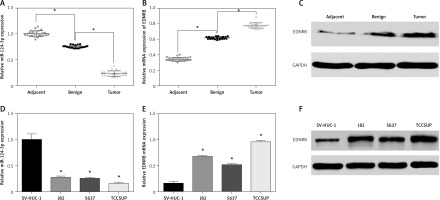
Firstly we evaluated the expression levels of miR-124-3p in tissue samples using qRT-PCR. The results showed that benign tissues had lower miR-124-3p expression than adjacent tissues and that tumor tissues had the lowest miR-124-3p expression among the 3 groups (p < 0.05, Figure 1 A). Conversely, EDNRB mRNA appeared to have up-regulated expression in BC tissues according to the qRT-PCR analysis (p < 0.05, Figure 1 B). The western blotting analysis which compared the EDNRB protein level in tissue samples also supported the previous result (Figure 1 C). Other results also suggested that BC cell lines had significantly lower miR-124-3p expression but higher EDNRB expression than normal cell lines (p < 0.05, Figures 1 D–F). The TCCSUP cell line which had the most significant variation from normal cell lines was selected to perform the following experiments.
Figure 1
Expression of miR-124-3p and EDNRB in BC tissues and cells. A – RT-PCR analysis of miR-124-3p in tissues. B – EDNRB mRNA detected with RT-PCR in tissues. C – Western blot analysis of EDNRB in tissues. D – miR-124-3p levels determined by RT-PCR in BC cell lines. E – RT-PCR analysis of EDNRB in BC cell lines. F – The EDNRB protein expression level in tissues as analyzed by Western blot
*p < 0.05.
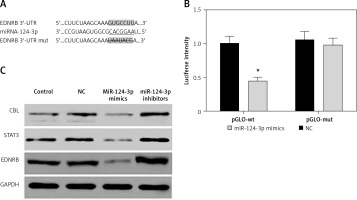
MiR-124-3p directly suppresses EDNRB expression in TCCSUP cells
As predicted by the Targetscan database, there is a targeted binding site for miR-124-3p in the EDNRB 3′-UTR region (Figure 2 A). The dual-luciferase reporter assay confirmed that cells expressing exogenous miR-124-3p displayed suppressed luciferase activity of the reporter plasmid carrying wild-type EDNRB 3′-UTR (p < 0.05). A similar suppressive effect did not take place when the binding sites were mutated (p > 0.05, Figure 2 B). MiR-214-3p has been reported to inhibit cancer cell proliferation by targeting Casitas B-lineage lymphoma (CBL), signal transducers and activators of transcription 3 (STAT3) [23, 24]. We examined the effect of miR-124-3p on CBL and STAT3 expression in BC cells as a comparison with previous studies. We further investigated the effect of miR-124-3p on EDNRB expression using western blotting. After the transfection of miR-124-3p mimics, endogenous CBL, STAT3 and EDNRB were remarkably down-regulated in TCCSUP cells, whereas their expression was up-regulated in cells transfected with miR-124-3p inhibitors (Figure 2 C). Therefore, miR-124-3p directly targets EDNRB in TCCSUP cells.
MiR-124-3p regulates the proliferation and apoptosis of BC cells by targeting EDNRB
As shown in Figures 3 A, B, the miR-124-3p mimics group had higher miR-124-3p expression and lower EDNRB expression, the EDNRB siRNA group had lower EDNRB expression and the miR mimics + EDNRB group had higher miR-124-3p expression (p < 0.05) than the control and NC groups. The MTS assay demonstrated that the 2 groups transfected with miR-124-3p mimics and EDNRB siRNAs had significantly inhibited cell viability compared to the control, NC and miR mimics + EDNRB groups (p < 0.05). Cells in the miR mimics + EDNRB group appeared to have similar viability compared to the control and NC groups (p > 0.05, Figure 3 C). The results of the colony formation assay were consistent with the results of the MTS. There was a significant reduction in the colony forming rate of cells in the miR-124-3p mimics and EDNRB siRNA groups compared to the control, NC and miR mimics + EDNRB groups (p < 0.05, Figure 3 D). Furthermore, the cell cycle assay demonstrated that overexpression of miR-124-3p or low expression of EDNRB can significantly induce G0/G1 phase arrest compared with control, NC and miR mimics + EDNRB groups (p < 0.05, Figure 3 E). In addition, the apoptosis assay revealed that the number of apoptotic cells dramatically increased in groups treated with miR-124-3p mimics and EDNRB siRNAs compared with the control, NC and miR mimics + EDNRB groups (p < 0.05, Figure 3 F). In conclusion, miR-124-3p exerted a suppressive function on tumor cell proliferation and induced cell apoptosis in BC cells by targeting EDNRB in vitro.
Figure 3
MiR-124-3p regulates proliferation and apoptosis of BC cells by targeting EDNRB. A – RT-PCR analysis of miR-124-3p expression. B – Western blot analyze EDNRB protein expression level. C – MTS Assay indicated that up-regulated expression of miR-124-3p or inhibition of EDNRB decreased the cell viability. D – Clonogenicity decreased in TCCSUP cells treated with miR-124-3p mimics or EDNRB siRNAs. E – The results of cell cycle analysis. F – Apoptosis rate increased after cells were treated with miR-124-3p mimics or EDNRB siRNAs. G – Tumor formation in nude mice
*P < 0.05 compared with control group.
Our in vivo experiments demonstrated that tumors induced with miR-124-3p overexpressed cells or EDNRB blocked cells had much slower growth compared with the control, NC and miR mimics + EDNRB groups (p < 0.05, Figure 3 G).
Discussion
A growing body of evidence suggests that miRNAs can contribute to bladder cancer development, progression and metastasis [25]. Deregulation of the endothelin axis has also been reported to occur in bladder cancer [26]. In our study, we found that miR-124-3p was expressed atypically in bladder cancer cells and played an important role in carcinogenesis. To verify the mechanism by which miR-124-3p exerts a suppressive effect on bladder cancer cells, we identified EDNRB as one of its targets. We then investigated its role in cell proliferation and apoptosis by manipulating its expression via cell transfection and have collected evidence to indicate that miR-124-3p restrains the malignancy of bladder cancer.
Recent studies have focused on the role that miRNAs plays in regulating the cellular processes of cancer cells [27]. As a result, numerous miRNAs including miR-124-3p have been studied in a variety of human malignant cancers. For instance, it was shown that miR-124-3p was significantly down-regulated in primary astrocytoma and that the overexpression of miR-124-3p can inhibit cell proliferation and invasion and stimulated cell apoptosis by directly targeting PIM1 [28]. In breast cancer, the inhibition of miR-124-3p enhanced the expression of Beclin-1 and can promote breast cancer cell progression [29]. In our study, we found that miR-124-3p expression was significantly reduced in both BC tissues and BC cell lines, especially in the TCCSUP cell line. This indicates that miR-124-3p can act as a tumor suppressor in BC. Furthermore, we found that the exogenous overexpression of miR-124-3p can suppress cell proliferation and induce cell apoptosis in the TCCSUP cell line. These results were also demonstrated in vivo via experimental tumor formation in nude mice.
EDNRB is one of the two endothelin G-protein-coupled transmembrane receptors found in non-vascular tissues [30]. Through the activation of EDNRB, endothelin-1 predominantly promotes endothelial cell proliferation and migration and can contribute to the process of angiogenesis in tumors [26]. This postulated role as an oncogene of endothelin-1 has been confirmed in various malignant cancers such as ovarian cancer, breast cancer and colon carcinoma [31–33]. In one previous study, overexpression of EDNRB was identified in 76.9% of 157 bladder cancer cases [22]. Therefore, as EDNRB also has high expression in both BC cell lines and tissues, this study indicated that it also has an oncogenic role in bladder cancer. However, the amount of direct research on EDNRB is limited. This has encouraged us to further investigate the role EDNRB plays in tumor progression. The results of the MTS and colony formation assay demonstrated that down-regulation of EDNRB can inhibit the cell proliferation of TCCSUP cells. This confirms its promoter role in the development and proliferation of BC cells. The flow cytometry assay also revealed that down-regulation of EDNRB can lead to an increase of BC cells in the G0/G1 phase and can induce cell apoptosis. Together, these results indicate that EDNRB participates in the tumorigenesis and progression of BC including cell proliferation and apoptosis.
Subsequently, we explored the specific relationship between miR-124-3p and EDNRB to further explore the underlying mechanisms involved in the suppressive process initiated by miR-124-3p. A bioinformatics analysis indicated that EDNRB was one of miR-124-3p’s targets. This was also confirmed by the luciferase reporter assay. Furthermore, we found that up-regulation of miR-124-3p can effectively inhibit the expression of EDNRB. This demonstrated that EDNRB was negatively affected by miR-124-3p.
In conclusion, the present study revealed that miR-124-3p exerted a remarkable suppressive function in bladder cancer. We also found that the up-regulation of miR-124-3p can suppress BC cell growth and induce cell apoptosis (in vitro and in vivo) by directly targeting EDNRB. Our study was the first to investigate the relationship between miR-124-3p and EDNRB in BC. We hope to provide new insight into the molecular mechanisms of BC progression and outline a novel treatment for BC. However, there are many miRNAs which target different genes. Therefore, it is possible that the results of our study may be partial and biased, and further research is still required.