Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
PULMONOLOGY / BASIC RESEARCH
The novel lncRNA PTTG3P is downregulated and predicts poor prognosis in non-small cell lung cancer
1
Department of Thoracic and Cardiovascular Surgery, Nantong First People’s Hospital, Nantong, Jiangsu Province, China
2
Division of Geriatrics, Tongji Hospital, School of Medicine, Tongji University, Shanghai, China
3
Department of Respiratory Medicine, Shanghai General Hospital, Shanghai Jiaotong University, Shanghai, China
Submission date: 2018-09-28
Final revision date: 2018-12-24
Acceptance date: 2019-01-04
Online publication date: 2020-03-09
Publication date: 2020-05-26
Arch Med Sci 2020;16(4):931-940
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Lung cancer is the leading cause of cancer-associated mortality worldwide. Recently, long non-coding RNAs (lncRNAs) have been studied as key regulators in some biological processes. Of note, the molecular mechanism and prognostic value of lncRNAs in non-small cell lung cancer (NSCLC) have largely remained unclear.
Material and methods:
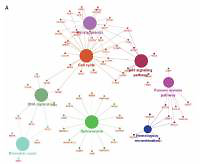
In this study, we compared the PTTG3P expression levels between lung cancer and normal lung samples by analyzing 5 public datasets (GSE18842, GSE19804, GSE27262, GSE30219, and GSE19188). Next, pentose phosphate pathway and co-expression networks were constructed to identify key targets of lncRNA PTTG3P. Furthermore, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed to explore the potential roles of lncRNA PTTG3P. Moreover, we constructed PTTG3P-mediated ceRNA networks in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC).
Results:
In the present study, our analysis showed that PTTG3P expression was higher in high T stage LUAD and LUSC samples, as well as high N stage NSCLC tissues. Of note, we found that higher PTTG3P expression is correlated with shorter survival time in NSCLC patients by analyzing Kaplan-Meier plotter datasets. We found that PTTG3P was significantly associated with NSCLC cell proliferation regulation by affecting a series of cell cycle related biological processes.
Conclusions:
Bioinformatics analysis showed that PTTG3P was associated with NSCLC cell proliferation. These results suggested that PTTG3P could serve as a new therapeutic and prognostic target for NSCLC.
Lung cancer is the leading cause of cancer-associated mortality worldwide. Recently, long non-coding RNAs (lncRNAs) have been studied as key regulators in some biological processes. Of note, the molecular mechanism and prognostic value of lncRNAs in non-small cell lung cancer (NSCLC) have largely remained unclear.
Material and methods:
In this study, we compared the PTTG3P expression levels between lung cancer and normal lung samples by analyzing 5 public datasets (GSE18842, GSE19804, GSE27262, GSE30219, and GSE19188). Next, pentose phosphate pathway and co-expression networks were constructed to identify key targets of lncRNA PTTG3P. Furthermore, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed to explore the potential roles of lncRNA PTTG3P. Moreover, we constructed PTTG3P-mediated ceRNA networks in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC).
Results:
In the present study, our analysis showed that PTTG3P expression was higher in high T stage LUAD and LUSC samples, as well as high N stage NSCLC tissues. Of note, we found that higher PTTG3P expression is correlated with shorter survival time in NSCLC patients by analyzing Kaplan-Meier plotter datasets. We found that PTTG3P was significantly associated with NSCLC cell proliferation regulation by affecting a series of cell cycle related biological processes.
Conclusions:
Bioinformatics analysis showed that PTTG3P was associated with NSCLC cell proliferation. These results suggested that PTTG3P could serve as a new therapeutic and prognostic target for NSCLC.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.