Introduction
Many risk factors have been suggested to predict coronary heart disease (CHD) [1–3]. The Framingham Risk Score [4], as well as the European SCORE [1], are the essential clinical tools for assessing the risk of cardiovascular events. The widely used SCORE system was introduced in the 2003 ESC guideline based on the results of 12 European cohort studies which took place between 1967 and 1991 [5]. Because of the timeline of the studies and the available data, only traditional risk factors were included in this scoring system; consequently, the system lacks certain crucial aspects, inter alia, the presence of diabetes or the results of novel studies dealing with inflammatory aspects of atherosclerosis, genetic parameters and respectively different types of familiar hypercholesterolemia. Delays and longer waiting lists caused by the inadequate selection of patients undergoing selective coronarographies, either due to the unnecessary examinations of low-probability patients or significant economic burdens suffered by the healthcare system, result in the late or even no diagnosis of the coronary artery disease. This so-called “over-diagnosing” is not associated with better prognosis; it merely highlights the need for optimisation of the examination process.
In the era of big data, available patient registries [6] and high computing power, it should be possible to appraise multiple parameters and personalised aspects based on the patient and the available data also with respect to the cardiovascular risk and the risk stratification of positive coronarography findings. The principal goal of the presented KSC MR study (method paper) is an attempt to establish novel approach in cardiovascular prevention based not on statistical methods but more personalised medicine supported by use of machine learning.
Machine learning in medicine
Although machine learning is not new to research, it is currently receiving quite a lot of attention. In recent years, there has been tremendous progress in the interplay between medicine and machine learning methods as manifested in data mining. Machine learning consists of various methods, the use of which ensures obtaining the desired results from analysed data. These methods are divided into supervised and unsupervised based on the algorithm used, which reflects the degree of control by the well-known classification of the analysed data into a category. Machine learning is used as part of artificial intelligence in many areas of human life, including healthcare and medical research. The medical industry is characterised by a huge increase in data and information describing the health status of patients, inventory descriptions, economic aspects of the operation of medical facilities, etc. Medical data, increasingly available in electronic form, are used for more effective healthcare provision, cost savings, or risk reduction in certain examinations. For example, the McKinsey Global Institute estimates that linking large data from the medical and pharmaceutical field to machine learning can annually save up to $ 100 billion in the US healthcare system [7]. In addition to the economic benefit, this fact has a positive impact on the patient, mainly in the form of primary and secondary disease prevention. The most widespread purpose of data mining in medicine is to help diagnose (identify) illnesses (e.g. breast cancer detection studies, classification of patients with heart failure as preserved or decreased left ventricular ejection fraction, etc.) [8]. The wide use of data mining is also reflected in the identification of various risk factors for certain diseases [9], for example, in the prediction of cholesterol levels in patients with myocardial infarction [10] or the mining of association rules for early prediction and risk of cardiovascular diseases [11]. Machine learning offers several types of methods to achieve a particular goal of the analysis. To a large extent, decision trees, which are used for ease of understanding of the patient classification, are easily interpretable (e.g. in the form of rules). On the other hand, artificial neural networks (now increasingly used in so-called deep learning) often provide a very good prediction or classification result (in relation to the purpose of the goal), but it is not clear from the result how it arrives at a conclusion, i.e. their explainability is very low. In this project, we will pay special attention also to the explainability aspect of the produced machine learning models which will be available to the cardiologists in the form of a decision support system with suitable visualisations and descriptions.
The Kosice Selective Coronarography Multiple Risk Study is aimed at preparing a predictive model for the identification of potentially risky patients and subsequently for this group of patients to indicate coronary angiography based on a model with a significant number of subjects under investigation. It also aims to determine the importance of the parallel presence of several risk factors for atherosclerosis and to gauge their effect on cardiovascular risk profile. We also foresee a deeper understanding of the interdependence of the various degrees of CVD disease occurrence as well as the understanding of individual factor combinations in patients where we anticipate an increased cardiovascular risk that is not confirmed just by invasive coronary angiography.
Material and methods
Patient population
The KSC MR study aims to enrol prospectively approximately 5,000 patients (Figure 1) aged 18 and above, without an upper limit, who will be admitted to the associated hospital. The monitored population will be male and female subjects referred for coronary angiography (CAG) due to suspected atherosclerosis of coronary arteries, but without a history of previously documented coronary artery disease (either acute or chronic coronary syndromes) – see complete exclusion criteria for atherosclerotic cardiovascular disease (ASCVD) in cardiovascular (CV) risk categories in 2019 ESC/EAS guidelines for the management of dyslipidaemias [1]. However, there is a whole range of different factors that have different levels of evidence and the clinical significance has not yet been clearly established. Our ambition is to either support such factors or contribute to their development as clinically relevant in deciding on the indication of CAG [12]. Coronary angiography will be performed in the East Slovak Institute of Cardiovascular Diseases in Kosice, Slovakia. The population will be selected in a way that ensures complexity for machine learning. All patients will be asked to sign an informed consent agreement in order to participate in this research project and their data will be anonymised.
Figure 1
Design of the KSC MR study with prospective and retrospective arm and groups of patients according to the SCORE and classification of CAG findings
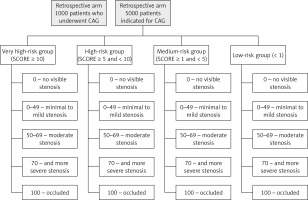
Patients will be divided into 4 groups according to the determined SCORE risk: very high-risk group (SCORE ≥ 10); high-risk group (≥ 5 and < 10) ; medium-risk group (≥ 1 and < 5) and low-risk group < 1. The high-risk countries’ SCORE system will be used because the patients will be selected from the Slovak population. Irrespective of risk based on SCORE calculation all 4 groups of patients included in the study will undergo coronary angiography (presence of medical indication). In accordance with the results of coronary angiography, findings will be classified as follows: 0 – no visible stenosis; 0–49 – minimal to mild stenosis; 50–69 – moderate stenosis; 70 and more – severe stenosis; 100 – occluded, based on the following publication [13].
Exclusion criteria for participation in this study will be: presence of coronary artery disease or other documented atherosclerotic cardiovascular disease (ASCVD) or significant atherosclerotic changes documented by previous computed tomography (CT) scan or carotid ultrasound [1] and/or diabetes mellitus, acute exacerbations of chronic inflammatory and/or autoimmune diseases (inflammatory bowel disease, rheumatoid arthritis, systemic lupus erythematosus, etc.), endo/myo/pericarditis, patients with malignant tumours (either under treatment or diagnosed a short time ago), inability to fill in the provided questionnaires, and lack of signed informed consent.
Study design
We will perform two types of studies, a prospective as well as a retrospective study (Figure 1). First, a retrospective study will be performed during the time when data from the prospective study are available. From the patients’ databases in the associated hospital, data of individuals (1000 available patients with accessible data in the last 4 years) who underwent CAG will be obtained. At the same time, all available clinical results (biochemical, haematological, as well as results of the clinical examination techniques) will be digitalised or transferred by the dedicated software to the anonymised database. The retrospective study enables us to analyse different types of machine learning models, both descriptive and predictive, in light of their suitability for the project goals. Those models can also be tested for the new data continuously coming from the prospective phase. The properly tested and optimised learning models will be further applied in the prospective phase with more available parameters and higher numbers of included patients.
In the prospective arm of the study, all patients admitted for CAG will be offered entry to this project. Before conducting CAG, we will collect from all patients anamnestic data and current clinical status information (physical examination, personal and family history and other source data provided by GPs and other physicians). In this step, patients will also be assigned to specific examinations such as standard 12-lead ECG, echocardiography or even liver FibroScan (transient elastography) for detection and determination of the severity of non-alcoholic fatty liver disease (NAFLD). We will also conduct dental examinations for assessment of the plaque/gingival/community periodontal index and the dental corrosion index and erosive changes and overall dental health. This step also entails taking biological material for further processing – biochemical determination of lipid profile, total cholesterol, high-density lipoprotein cholecterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), TAG and individual fractions of LDL-C and HDL-C using the Lipoprint method, PCSK9, high-sensitivity C-reative protein (hsCRP), hepatitis B virus (HBV), hepatitis C virus (HCV), human immunodeficiency virus (HIV), cytomegalovirus (CMV) and screening of infectious diseases as well as whole-genome sequencing and selected miRNA analysis along with the control group.
Group and data size
The amount of data required for machine learning depends on many factors, but mainly on the complexity of the problem (i.e. the unknown underlying function that best relates input variables to the output variable), and the complexity of the learning algorithm (i.e. the machine learning algorithm used to inductively learn the unknown underlying function from given examples). In both of these aspects we assume a high level of complexity, which implies a larger amount of data needed for the learning process.
Firstly, many studies focused on cardiovascular risk have evidenced quite a large number of relevant parameters and some more may still not be known or evidenced well enough. This fact implies high complexity of the underlying function.
Secondly, we plan to use different machine learning algorithms ranging from less complex ones (such as logistic regression) up to very complex ones (deep neural networks). Therefore, we need to assume also highly complex models which aim to properly cover also less frequent groups of patients, e.g. those with a low value of the calculated cardiovascular risk score (e.g. in terms of SCORE) on one hand, but with serious coronary findings on the other hand. For the retrospective group the number 1000 has been set as a statistically representative sample of the Slovak population. But for the prospective group, a much larger number will be needed. One reason is that for the retrospective group we have significantly fewer parameters available (about 60) than for the prospective group (from 2 up to 3 times more). Statistical analysis is not required for this kind of project because no comparative values will be analysed. We are not going to identify or correlate any of the measured values/velocities, and no correlation is going to be used to prove our concept for different risk factors/risk groups of patients. The aim of this trial is to use specific algorithms for machine learning or neural networks to identify logical or non-logical pathways for patient identification or patient risk identification for a future CV major adverse cardiovascular events (MACE) event based on standard of care values and information such as lab reports, angiography reports, echocardiography reports, etc.
Processing and availability of data, machine learning protocols
A team of researchers from the Data Science research group (Department of Cybernetics and Artificial Intelligence, Technical University of Košice, Slovakia) will be responsible for the machine learning part of the project. All the data will be available in anonymised form for the analytical process. The data analysts will not know which record relates to which specific patient. The anonymization will take place at the level of the medical workplace during the extraction of data from medical records. The data analysts will use relevant data samples on selected local computers with all the necessary security safeguards/guarantees. Public cloud services will not be used.
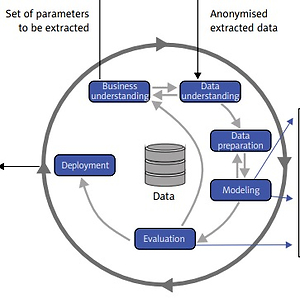
Data extraction processes for data coming from EHRs are basically the same for both the retrospective and the prospective study, but the number of parameters will be much higher for the prospective study. In addition, qualitative data from the interviews will be processed as well for the prospective study. The data extraction process has already been implemented and tested; the details can be found in [14]. The result are anonymized data available in the form of a table, which is further processed by means of the CRISP-DM methodology (Figure 2) [15]. When applying this methodology in the medical domain, it is extremely important that the data analyst closely cooperates with the domain experts (i.e. cardiologists in this case). Main tasks of the data analyst and domain expert in particular phases of the CRISP-DM methodology are outlined in Figure 3 [16]. We successfully used this methodology in our previous studies in various medical applications [17, 18].
Figure 2
CRISP-DM methodology [15] will be used for the data analytics part of the project. Machine learning and artificial intelligence approaches are applied in the modeling phase. Both predictive and descriptive models will be trained and evaluated (CRISP DM – Cross Industry Standard Process for Data Mining; EHR – electronic health record; kNN – k-nearest neighbors; CBR – case-based reasoning)
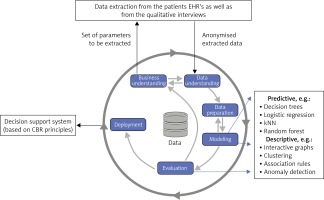
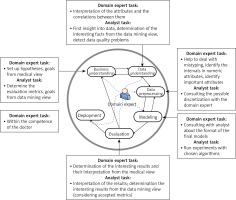
Figure 3
CRISP-DM methodology with highlighted tasks of the domain expert (cardiologist) and data analyst (machine learning expert) [16]
In the modeling phase of the CRISP/DM methodology, we will apply various machine learning algorithms, which can be divided into two main groups. First, predictive models will be trained by means of algorithms producing easy interpretable models such as decision trees, logistic regression or SVM. But we plan to experiment also with neural network models especially when image data are available. For (possibly deep) neural networks or random forests and other less interpretable models we will provide methods to support their explainability, which is crucial in the medical applications. The second group of modelling approaches will be descriptive ones. We will provide a description of extracted summarized data in the form of interactive graphs of various types and plan to use different machine learning approaches to clustering, association rules mining and anomaly detection, which may be interesting models for this study.
Finally, suitable deployment of the resulting models will be provided in the form of a decision support system for cardiologists. The first prototype of this system has already been developed and based on the data from the retrospective study tested by a cardiologist. The design and first results will be published in [19]. This first prototype is based on the case-based reasoning principles [20], making use mainly of the k-NN (k-nearest neighbors) classifier. But e.g. for one of the visualizations the PCA (principal component analysis) method has been implemented.
Results and Discussion
This study-based CHD risk stratification, unlike the most commonly used scoring system in preventive cardiology for CV risk calculation, aims to assess the full spectrum of data and parameters for each patient enrolled (starting with thoroughly analysed and correctly interpreted anamnestic data, through the broad spectrum of laboratory and other examinations methods). Our ambition is, therefore, to indicate individual diagnostic methods in a timely and correct manner as well as to preclude their unnecessary use where they are not indicated.
We believe that by using a combined approach of personalised medicine and machine learning, we could be able to contribute partially to the improvement of the morbidity and mortality situation in CHD. Each patient is a unique being and therefore deserves a unique approach. Without extensive use of modern data (computing) technologies, this would probably hardly be an achievable goal. The project will provide insights into the connection and significance of individual risk factors at different levels previously studied separately due to the various limitations of research projects as well as the inappropriate use of the latest machine learning options through the analysis of large patient databases in preventive cardiology in the past. We expect that better CV risk management can be reached not only by traditional risk factors’ identification and their possible elimination or treatment. Our hypothesis is that substantial overuse of several diagnostic methods and laboratory techniques could be diminished and more precise selection of investigation methods could be achieved in the future by more extensive use of personalised medicine in combination with methods of machine learning.
Through the combination of medicine and information technologies, this project will help to evaluate the weight of individual risk factors in the occurrence of detectable coronary changes/increased CV risk, the development of evaluation and predictive models which will also give preference for prevention, as well as therapeutic activities in the area of CV disease. We suggest that the study will give rise to an additional system based on the principles of personalised medicine and all available patient data.