Introduction
The Demodex mite was first reported by Gustav Simon, a German dermatologist examining sebaceous follicle samples from an individual with acne lesions [1]. Simon first identified Demodex folliculorum in 1842, but it was not until 1963 that Demodex brevis was identified as a separate species [2].
Demodex mites are the most common human ectoparasites [3]. Demodex species can be found on all skin types across a broad geographical range [2]. Demodex mites reside in the pilosebaceous units of the skin, with their heads directed at the bottom of the hair follicle, where most sebum can be found [4]. Demodex feed on epithelial and glandular cells as well as sebum typically secreted by active pilosebaceous units [5, 6].
Demodex folliculorum is a more common mite, possibly due to it inhabiting the upper end of the sebaceous unit and so making it easier to spread by direct contact [4]. This saprophytic mite colonizes the facial region commensally with D. brevis residing deep within the sebaceous gland of the skin and the meibomian gland of the eye [7, 8].
It is believed that Demodex mites are first transmitted during nursing as Demodex mites have been isolated from the nipple [9]. As children mature, the sebaceous unit develops, creating a suitable habitat for Demodex colonization, which further improves as young adults produce sebum during puberty and change its composition [10]. Demodex densities increase with age, with almost 95% infestation found in individuals over the age of 71 [9].
As a result of Demodex feeding on sebum and epithelial cells, particularly around the nose, as it is rich in sebum, this may cause micro-abrasions to the skin surface [11]. This can commonly lead to epithelial hyperplasia on the face or even the eyelid margin of rosacea patients and blepharitis patients [11]. It has been suggested that the sebum composition rather than its volume is preferred by the mite and can determine Demodex densities [12]. Demodicosis is the infestation of Demodex mites on the face, whereby a minimum of 5 mites/cm2 exist and induce symptoms such as redness of the skin (erythema), telangiectasia, itching, heat, and scaling [8].
In 1971 the skin surface biopsy was described to detect D. folliculorum presence on the skin. However, it was not until 1993 that the concept of assessing the Demodex density was first introduced [13–15]. This quickly led to the development and utilization of the standardized skin surface biopsy (SSSB) method on the stratum corneum, immediately followed by a second deeper skin surface biopsy as outlined by Forton and De Maertelaer [15]. By combining these two consecutive samples’ counts, an improved Demodex density could be found, proving this method as a useful diagnostic tool. The SSSB is a quick and reproducible procedure, tolerated well by patients as it is minimally invasive. It is also a cheap and straightforward diagnostic tool. The two consecutive SSSBs reached 89.3% sensitivity, compared to 55% with the use of just one biopsy [15].
A different modification of SSSB was proposed by Arasiewicz et al. In their approach, a hypoallergenic, sticking plaster is pressed onto the nose, chin, cheeks, and forehead. These are then removed after a night, followed by a microscopic analysis [16].
However, these microscopic methods present certain disadvantages. Performing a double SSSB is time-consuming, with sensitivities below 90%. The use of cyanoacrylic glue might also scare the patients. Thus, genetic approaches have been under examination. They give an opportunity to significantly increase sensitivity and provide additional information from the genetic material of the mite, the host, and the surrounding microbiota.
However, it has not been verified if non-invasively collected sebum can be successfully used by Next-Generation Sequencing (NGS) in Demodex analysis.
Aim
The purpose of this preliminary study was to assess whether non-invasively collected sebum samples of patients suspected of rosacea or demodicosis are suitable for NGS DNA Demodex analysis.
Material and methods
Patients
Suspicion of seborrheic dermatitis (criteria of the Polish Dermatological Society [17]) or rosacea (criteria of the National Rosacea Society [18]) manifesting themselves by facial skin inflammation was the inclusion criterion. Exclusion criteria involved age of under 18 and treatment within last 4 weeks. One person was free of symptoms and served as a negative control. The entire group consisted of 21 people: 20 males, 1 female, age range: 33–83, median age: 58. All patients gave written consent. The study was approved by the Bioethical Committee of the Poznan University of Medical Sciences.
Sebum collection
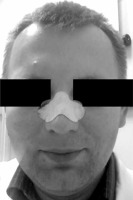
Sebum and epidermal debris were collected with the use of Deep Cleansing Face Pore Strips (Beauty Formulas, UK). Nasal dorsum was moisturized with PBS, and a strip was applied to it for 10 min (Figure 1). In most cases, the strips were applied before the patients had their face cleaned or cream applied. Purposefully, the skin was not pinched to minimize the level of invasion, even though pinching could have yielded more sebum. Then, the strip was removed, put into a plastic zip bag, and immediately frozen at –20°C. The freezing step allowed the adhered sebum to harden and made it possible to be scratched off from the strip with a scalpel. The peeled debris were put into a 2.0 ml Eppendorf tube with lysis buffer.
DNA isolation
Due to minute amounts of input material and high fat content, DNA isolation proved to be challenging. Best results were achieved with the use of a NucleoSpin Food DNA isolation kit (Macherey-Nagel). The procedure was carried out according to the manual with slight modifications in that additional vortexing was applied during initial Proteinase K and lysis buffer incubation followed by centrifugation at 11000 xg. DNA was eluted in only 30 µl elution buffer (5 mM Tris/HCl, pH 8.5) to yield a higher concentration. The DNA concentration was measured with Qubit fluorometer and Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). Isolated DNA was stored at –20°C prior to further processing.
Library preparation and validation
1.0 ng of each DNA sample was used as input for library preparation with the use of Nextera® XT DNA Sample Preparation Kit (Illumina), according to the user manual. Libraries were indexed with a dual-indexing strategy using Nextera® XT Index Kit (Illumina) that enabled multiplexing of the samples. The distribution of fragment length and concentration of final libraries was assessed by means of on-chip electrophoresis using Agilent High Sensitivity DNA Kit and Agilent Bioanalyzer 2100 System (Agilent). Each library was diluted to 2 nM concentration, an equimolar pool of all libraries was prepared, denatured, and diluted to the final 8 pM concentration [19].
NGS sequencing
The diluted pool of libraries was sequenced in pair-end, 2 × 150 cycles mode, with the use of MiSeq® Reagent Kit v2 (300 cycles) and MiSeq® System (Illumina), according to user manuals. 5% of PhiX Control (Illumina) was spiked in samples to obtain run quality metrics.
Bioinformatic analysis
The following analyses were performed using the Galaxy Web Portal at usegalaxy.org [20], apart from the NCBI BLASTn suite [21].
Elimination of human reads
Since the data from two runs in MiSeq® System paired-end (PE) sequencing are stored as individual files, a list of data-set pairs was created. These pairs were then mapped against the Homo sapiens hg38 reference genome to eliminate human DNA reads using Bowtie2 v.2.3.4 with default settings [22, 23]. Read pairs that did not align concordantly to the human genome were processed further.
Mapping against Demodex mitochondria
Since the full genomic sequences of both Demodex folliculorum and brevis are not yet available, the unaligned reads were mapped against their known mitochondrial genomes (NC_026102.1 and NC_026101.1, respectively [24]) using Bowtie2. Mapped results were then filtered and converted into FASTA format with BAM Tools (Galaxy Version 2.4.0) [25].
The FASTA files were used to perform NCBI BLASTn searches as the final check of the results (‘Somewhat similar sequences’ mode, ‘Nucleotide collection (nr/nt)’ database, ‘Low complexity regions’ unchecked) [21]. Reads were considered positive if the hit with the lowest e-value was referring to the appropriate Demodex species. The BLASTn confirmation step was omitted for samples with more than 100 PE reads mapped.
Results
The results of the DNA isolation are provided in Table 1. Unfortunately, there was not enough facial material yielded from some patients, resulting in unsuccessful DNA isolation. A total of 5/21 (23.8%) isolation attempts failed. All of these happened in the first half of the collected samples. During this time we established two additional sample entry criteria: 1) the sample must be collected before any dermatological treatment starts, 2) frozen sebum must be visible on a strip before scratching it with a scalpel.
Table 1
Results
| Patient*/sex | Age | DNA concentration [ng/µl] | Microscopic analysis | Paired-end reads total | Non-human PE reads | D. folliculorum mapped PE reads | BLASTn D. folliculorum confirmed PE reads | D. brevis mapped PE reads | BLASTn D. brevis confirmed PE reads |
|---|---|---|---|---|---|---|---|---|---|
| 1/M | 58 | 6.30 | Negative | 638554 | 72019 (11.28%) | 0 | – | 0 | – |
| 4/M | 61 | 75.0 | Negative | 605509 | 70936 (11.72%) | 0 | – | 0 | – |
| 6/F | 61 | 0.42 | Negative | 519298 | 81469 (15.69%) | 2 | 2 | 4 | 4 |
| 7/M | 65 | 0.15 | Negative | 963428 | 114277 (11.86%) | 31 | 31 | 0 | – |
| 8/M | 40 | 33.3 | Negative | 672734 | 79421 (11.81%) | 0 | – | 0 | – |
| 11/M | 33 | 0.45 | Positive | 728772 | 84344 (11.57%) | 0 | – | 0 | – |
| 12/M | 66 | 0.16 | Positive | 970263 | 443816 (45.74%) | 59 + 1 SE | 59 + 1 SE | 11 + 1 SE** | 11 |
| 13/M | 55 | 0.14 | Positive | 764057 | 144228 (18.88%) | 260 + 2 SE | Not assessed | 1** | 0 |
| 14/M | 41 | 1.05 | Positive | 690920 | 110420 (15.98%) | 1 + 1 SE | 1 + 1 SE | 0 | – |
| 15/M | 34 | 0.24 | Negative | 671291 | 121220 (18.06%) | 0 | – | 0 | – |
| 16/M | 58 | 0.22 | Negative | 627240 | 73727 (11.75%) | 14 + 1 SE | 14 + 1 SE | 0 | – |
| 17/M | 54 | 0.25 | Positive | 638441 | 72116 (11.30%) | 100 + 1 SE | Not assessed | 0 | – |
| 18/M | 73 | 0.78 | Negative | 615519 | 241721 (39.27%) | 4 | 4 | 0 | – |
| 19/M | 37 | 1.93 | Negative | 671320 | 77000 (11.47%) | 0 | – | 0 | – |
| 20/M | 83 | 0.41 | Positive | 1132927 | 216371 (19.10%) | 25 | 25 | 1 | 1 |
| 21/M | 76 | 0.20 | Positive | 342636 | 253606 (74.02%) | 1 | 1 | 0 | – |
| N studied | 16 | 10/16 | 10/16 | 4/16 | 3/16 | ||||
| Min. | 33 | 0.14 | 342636 | 70936 (11.28%) | |||||
| Max | 83 | 75 | 1132927 | 443816 (74.02%) | |||||
| Mean | 55.9 | 7.56 | 703307 | 141043 (21.2%) | |||||
| Median | 58 | 0.42 | 671306 | 97382 (13.8%) | |||||
Run quality
Overall quality of the sequencing run was high (cluster PF = 91%, > Q30 = 95%; error rate = 0.6) and over 22 million passing filter sample reads were generated (0.7–2.2 million per sample).
Elimination of human reads
Firstly, both the default and sensitive-local options in Bowtie2 were run to compare the two modes for the best results. Initial analysis showed that the surplus hits generated by the --sensitive-local option are in fact human reads that were not removed in the primary elimination of human reads or hits, yielding no results under BLASTn. It was therefore decided to use the default options of Bowtie2.
As expected, most of the DNA was indeed human. The summary of human DNA elimination is provided in Table 1.
Demodex detection
No statistically significant correlation was observed between the initial DNA concentration and the number of paired-end reads yielded (Spearman rs = –0.28, p = 0.29).
Out of the 7 patients that were previously confirmed positive by microscopic analysis, 6 were also found positive by NGS. One patient (no. 11) was found negative by NGS and positive by microscopy. On the other hand, additional 4 patients were found positive by NGS, being negative by microscopy, adding to a total of ten. Thus, the NGS approach showed superior sensitivity compared to light microscopy (63% and 44%, respectively).
Interestingly, in 3 patients, both Demodex species were detected by NGS. One of these patients was negative in light microscopy. It is worth mentioning that the BLASTn analysis was unambiguous in all 3 cases. This step seems crucial, however, since it pointed to 2 false positive hits. One of these was a SE read with no influence on the final results since other PE reads were present. The other was a single PE read for that patient (no. 13); thus, the status was changed from positive to negative only as far as Demodex brevis was concerned since this patient was strongly positive for Demodex folliculorum.
Discussion
Genomic, transcriptomic, and microbiotic analyses of human Demodex species are a challenge. The mites cannot be cultured in vitro but only kept alive for a few days [26, 27]. Several genetic analyses of Demodex mite have been carried out. For instance, Thoemmes et al. carried out a phylogenetic analysis on the 16S rRNA and the 18S rRNA genes to determine possible genetic diversities and the lineage history of the Demodex mite evolution [2]. A genetic variance between the two Demodex species has been highlighted as mites located at the eyelashes have a different CO1 mitochondrial gene compared to mites inhabiting the skin [28].
Until now, Demodex mites have been obtained from sebaceous glands of individuals by various techniques such as scraping the skin gently, cellophane tape, plucking in the case of eyelashes and eyebrows, and also by invasive methods, i.e. a biopsy [2]. Nevertheless, the mites are microscopic, thus difficult to handle, while the amount of the sample for any DNA or RNA analysis is far from desirable. It is the methodology of extraction that must be resolved to successfully diagnose patients with high sensitivity, not only qualitatively but quantitatively, allowing thorough scientific research to be done. Our extraction and DNA isolation methodology was acceptable as far as this research was concerned but obviously requires more insight and optimization. Perhaps, not pinching the skin to minimize the invasion level was a mistake since a visible sebum sample at this early stage was important for later DNA isolation success.
The authors are aware that at the moment the usage of NGS in the diagnostics of individual patients is not likely due to the overall cost of the procedure per patient. The procedure might be used in such a way in the future as the prices per sample decrease.
For the time being, genetic diagnostics of Demodex parasites can be aided by PCR and second-generation sequencing that are far cheaper per patient yet less informative. Worth mentioning is the ongoing research for the most suitable barcoding regions [29, 30].
However, we believe to have proven that it is now possible to study the Demodex genomics in individual people. Coupled with the microbiota analysis of both the outside and inside of mites inhabiting the skin, it opens new research possibilities of that pathogen.
Furthermore, we have been able to successfully identify and distinguish Demodex folliculorum from D. brevis in individual patients. Both human Demodex mites coexist commensally and share common characteristics but also have structural differences and habitual preferences. D. brevis is the smaller of the two species and is solitarily confined deep within the glands and ducts but is located in more widespread areas of the face such as the ear canals [9]. The two species reside in different ecological niches, thus might require different treatment approaches. The authors are also aware that relying on just one mitochondrial sequence per species as reference points is likely not the ideal situation as far as specificity is concerned. Our conclusion that the two Demodex species can be specifically distinguished from one another and from other mites is based on the assumption that any yet to be discovered mitochondrial diversity would not overlap. That is likely the case, but the possibility of a much smaller genetic distance is not entirely excluded. More sequences need to be deposited so that researchers can assign species to their sequences with more certainty. Worth mentioning is the outstanding research on the transcriptome of Demodex mites recently made by Hu et al. It paves the way for understanding the differences between the two human invading species and developing reliable treatment strategies [27, 31].
The causality of Demodex spp. in the etiopathogenesis of rosacea is a subject of an ongoing scientific debate. One of the most common hypotheses on the matter suggests it may act like a syringe by bringing bacteria and antigens deeper into the skin than normally present. This can be especially seen when mites die deep in pilosebaceous units releasing contents of their intestines. This hypothesis is supported by studies pointing to certain bacteria, such as Bacillus oleronius, whose proteins have been shown to elevate inflammatory response in vitro [3, 7, 11, 12, 32–34]. These studies show that further analyses on the host-mite-bacteria interactions in the etiopathogenesis of rosacea are necessary. NGS enables researchers to look for genetic traits of these bacteria and even study the entire microbiota inside or outside mites, both in vitro and in patients.