Introduction
Psoriasis is a chronic autoimmune disease characterized by thick, scaly plaques resulting from epidermal hyperplasia and immune cell infiltration. Its prevalence varies globally, and is influenced by genetic, ethnic, and environmental factors. Triggers such as stress, infections, seasonal changes, and medications like β-blockers contribute to disease onset and exacerbations [1]. The interplay between keratinocytes and immune cells plays a key role in its pathogenesis, with recent research highlighting keratinocytes as active participants in immune-mediated skin changes [2].
Central to psoriasis pathogenesis is the interleukin-23 (IL-23) – T helper cell 17 (Th17) axis. IL-23 activates the Tyk/Jak2 and STAT3 pathways, promoting Th17 cell proliferation and cytokine secretion, including IL-17, IL-21, and IL-22. These cytokines drive keratinocyte proliferation, angiogenesis, and immune infiltration. Tumor necrosis factor α (TNF-α) and interferon-γ (IFN-γ) further amplify inflammation by enhancing antigen presentation and inducing proinflammatory mediators. Together, these pathways establish a proinflammatory environment that perpetuates psoriatic lesions [3, 4].
The cutaneous microbiome of psoriatic patients is markedly altered, exhibiting reduced diversity and an overgrowth of opportunistic pathogens like Staphylococcus aureus, Streptococcus pyogenes, Candida albicans, and Malassezia. These pathogens contribute to disease flares by triggering immune responses and cytokine production. Variations in skin microbiota, including shifts in bacterial and fungal communities, suggest a role for microbiome-targeted therapies in managing psoriasis [5].
Polymorphisms in cytokine genes, including IL-23, are linked to immune dysfunction in various diseases. Understanding these polymorphisms and their haplotypes could provide deeper insights into their role in psoriasis severity and susceptibility [6].
Aim
This study aims to investigate the skin microbiome variations between psoriatic patients and healthy controls while examining the correlation of IL-23 and its gene single nucleotide polymorphisms (SNPs) with the presence and severity of psoriasis.
Material and methods
Study design and population
This case-control study included 80 participants recruited from the Dermatology, Venereology, and Andrology Outpatient Clinic at the Benha University. Ethical approval was obtained from the Research Ethics Committee, Faculty of Medicine, Benha University. Informed consent was provided by all participants before enrolment (Approval code: 41-4-2023).
Eligibility criteria
Inclusion criteria
Participants included both males and females aged 16 to 45 years, clinically diagnosed with plaque psoriasis, including those experiencing worsening of the existing lesions or the development of new lesions in the past month. Eligible patients must not have received psoriasis treatment in the preceding month and must have a Psoriasis Area and Severity Index (PASI) score of ≥ 18, covering mild, moderate, and severe cases (PASI < 10, 10–20, and > 20) [7]. Healthy controls with no history of psoriasis or autoimmune diseases, including familial history, were also included. Volunteers of all ages and genders were eligible to serve as controls.
Exclusion criteria
Exclusion criteria included patients with obesity; pregnant women or those using hormonal contraception; individuals who had received narrowband ultraviolet B irradiation or systemic treatment in the past month; or those who had undergone any psoriasis treatment. Additional exclusions were applied to patients with immune-mediated comorbidities, malignancies, systemic diseases, or a history of using topical or systemic antifungals or antibiotics within the past month.
Grouping
The 80 participants were divided into two equal groups. The Psoriasis Group consisted of plaque psoriasis patients whose diagnoses were based on clinical findings, dermoscopic assessment, and, if necessary, confirmed by histopathological studies. The Control Group included apparently healthy individuals matched for age and sex to the patient group.
Assessments
Participants provided detailed personal, medical, and family histories. Clinical examinations included a general physical and a focused dermatological assessment in well-lit conditions, evaluating lesions on the skin, mucous membranes, nails, and hair. Psoriasis vulgaris diagnosis was based on clinical presentation, dermoscopy, or biopsy when needed.
Disease activity was measured using the PASI score, which evaluates lesion severity and treatment response. PASI scores range from 0 to 72, with 5–10 indicating moderate disease and > 10 severe disease. The body is divided into four regions – head, upper extremities, trunk, and lower extremities – assessed for erythema, induration, and scaling, with a calculated formula integrating regional scores [7].
Dermoscopy and skin microbiome analysis
Dermoscopy
Dermoscopy confirms the diagnosis of psoriasis through characteristic findings, including the presence of dotted vessels – dilated, elongated, and convoluted capillaries – accompanied by a light red background and white superficial scales [8].
Skin microbiome detection
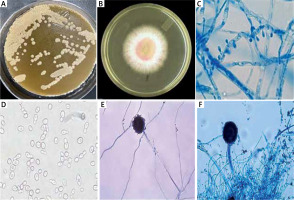
For microbiome analysis, scales were scraped from psoriasis lesions to identify bacterial and fungal organisms. Detection methods included microscopy, culture, biochemical reactions, and the use of specialized agar media (Figures 1 A–F).
Figure 1
Dermoscopic and microbiome analysis of psoriatic lesions, A – Malassezia furfur colonies on Sabouraud dextrose agar overlaid by olive oil, B – Dermatophytes growth on Sabouraud dextrose agar containing cyclohexamide, C – Dermatophytes, D – Candida albicans show germ tube formation, E – Aspergillus hyphae, F – Aspergillus head. This figure demonstrates the altered skin microbiome in psoriasis, highlighting increased fungal and bacterial colonization in psoriatic lesions.
Assessment of IL-23
Blood sample collection
Venous blood samples (2 ml) were collected from all participants. Samples were allowed to coagulate for 30 min before being centrifuged for 2 min. The serum was then stored at –80°C until analysis.
Cytokine measurement
Serum levels of IL-23 were quantified using a double-antibody sandwich enzyme-linked immunosorbent assay (ELISA) with commercially available kits specific for the p19 subunit of IL-23. All assays were conducted by technicians blinded to the clinical status of the participants.
ELISA procedure
A standard curve was generated by serially diluting the standard to concentrations of 96, 48, 24, 12, and 6 pg/ml.
For blank wells, only chromogen solutions A and B, and stop solution were added.
For standard wells, 50 μl of standard and Streptavidin-HRP were added.
For test wells, 40 μl of sample, IL-23 antibodies, and Streptavidin-HRP were added.
The plates were incubated at 37°C for 60 min with gentle shaking. Following incubation, the wells were washed, and chromogen solutions were added. The reaction was incubated for 10 min before being stopped with the addition of a stop solution. Optical density (OD) was measured at 450 nm within 15 min. Serum cytokine levels were calculated from the standard curve based on OD values.
Genotyping of IL-23
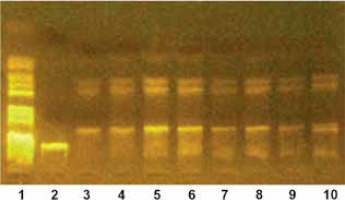
Genotyping of IL-23 SNPs was carried out using the polymerase chain reaction (PCR) technique followed by restriction fragment length polymorphism (RFLP) analysis. This method enables the detection of genetic variations by amplifying specific regions of DNA and then digesting them with restriction enzymes. The resulting DNA fragments were separated electrophoretically, providing insights into genetic variability (Figure 2).
Statistical analysis
Data analysis was performed using the Statistical Package for the Social Sciences (SPSS) software, version 25 (SPSS Inc., PASW Statistics for Windows, Version 25, Chicago: SPSS Inc.). Qualitative data were described in terms of numbers and percentages, while quantitative data were summarized using median and range for non-normally distributed variables and mean ± standard deviation (SD) for normally distributed variables. Normality of data distribution was assessed using the Kolmogorov-Smirnov test. The level of significance for statistical analysis was set at ≤ 0.05. To compare qualitative data between groups, the χ2 test, Fisher’s exact test, and Monte Carlo tests were applied, as appropriate. These statistical methods ensured robust and reliable comparison of variables to support the study’s conclusions.
Results
The comparison between patients with plaque psoriasis and the control group revealed no significant differences in gender, age, or occupation. Both groups had identical gender distributions, with 57.5% of males and 42.5% of females (p-value = 1). The mean age was similar, with 35.75 ±9.63 years in the psoriasis group and 35.98 ±9.43 years in the control group, and age ranges from 16 to 45 years in both groups (p-value = 0.961). Occupational analysis showed comparable patterns, with around half of each group being unemployed (50% in the psoriasis group and 57.5% in the control group), and the remaining participants were predominantly workers, with no significant differences (p-value = 0.737).
The median duration of psoriasis was 3.0 years, ranging from 0.08 to 12.0 years. The most commonly affected sites were the upper limbs (50.0%), followed by the lower limbs (42.5%), trunk (10.0%), and head (7.5%). A family history of psoriasis was reported in 22.5% of patients, while the majority (77.5%) had no family history. Based on PASI scores, 72.5% of patients had mild disease (< 10), 25.0% had moderate disease (10–20), and 2.5% had severe disease (> 20), with a median PASI score of 5.05 (range: 0.70–20.70).
Patients had significantly higher rates of fungal growth alone (20.0% vs. 2.5%, p = 0.029) and mixed bacterial and fungal growth (55.0% vs. 15.0%, p < 0.001) than controls, while bacterial growth alone was higher in controls (47.5% vs. 25.0%, p = 0.036). All patients showed microbial growth, compared to 35.0% of controls with no growth (p < 0.001). Staphylococcus aureus (42.5% vs. 10.0%, p = 0.001) and Streptococcus pyogenes (25.0% vs. 2.5%, p = 0.003) were more common in patients, whereas Staphylococcus epidermidis was more frequent in controls (32.5% vs. 7.5%, p = 0.005). No significant differences were found for Staphylococcus saprophyticus (2.5% vs. 5.0%, p = 1.000) or Corynebacterium acne (2.5% vs. 12.5%, p = 0.201). Among fungi, Candida albicans (37.5% vs. 5.0%, p < 0.001), Aspergillus niger (55.0% vs. 10.0%, p < 0.001), and Malassezia (22.5% vs. 2.5%, p = 0.007) were significantly more prevalent in patients (Table 1).
Table 1
Bacterial and fungal growth, and culture in the studied groups
| Parameter | Patients (n = 40) | Controls (n = 40) | Test (χ2) | P-value |
|---|---|---|---|---|
| n (%) | n (%) | |||
| Any growth | ||||
| No growth | 0 (0.0%) | 14 (35.0%) | 16.970 | < 0.001* |
| Bacterial growth alone | 10 (25.0) | 19 (47.5%) | 4.381 | 0.036* |
| Fungal growth alone | 8 (20.0%) | 1 (2.5%) | 6.135 | 0.029* |
| Both bacterial and fungal growths | 22 (55.0%) | 6 (15.0%) | 14.066 | < 0.001* |
| Multiple growth | ||||
| Single | 18 (45.0%) | 20 (76.9%) | 6.574 | 0.01* |
| Mixed | 22 (55.0%) | 6 (23.1%) | ||
| Bacterial growth | ||||
| Negative | 8 (20.0%) | 15 (37.5%) | 2.99 | 0.084 |
| Positive | 32 (80.0%) | 25 (62.5%) | ||
| Fungal growth | ||||
| Negative | 10 (25.0%) | 33 (82.5%) | 26.6 | < 0.001* |
| Positive | 30 (75.0%) | 7 (17.5%) | ||
| Bacterial culture | ||||
| Staphylococcus aureus | 17 (42.5%) | 4 (10.0) | 10.912* | 0.001* |
| Staphylococcus epidermis | 3 (7.5%) | 13 (32.5) | 7.813* | 0.005* |
| Staphylococcus saprophyticus | 1 (2.5%) | 2 (5.0) | 0.346 | 1.000 |
| Streptococcus pyogenes | 10 (25.0%) | 1 (2.5) | 8.538* | 0.003* |
| Corynebacterium acne | 1 (2.5%) | 5 (12.5) | 2.883 | 0.201 |
| Fungal culture | ||||
| Candida albicans | 15 (37.5%) | 2 (5.0%) | 12.624 | < 0.001* |
| Aspergillus niger | 22 (55.0%) | 4 (10.0%) | 18.462 | < 0.001* |
| Malassezia | 9 (22.5%) | 1 (2.5%) | 7.314 | 0.007* |
The comparison of IL-23 serum levels between patients and the control group revealed significantly lower levels in patients (157.2 ±214.8) compared to controls (252.7 ±308.2) (p < 0.001). The median IL-23 serum level was also lower in the patient group (58.83) compared to the control group (134.1), with the statistical analysis confirming a significant difference between the two groups.
The Hardy-Weinberg equation was applied to calculate the expected genotype counts for all studied SNPs, and the results revealed that both controls and plaque psoriasis groups were in Hardy-Weinberg equilibrium. No significant differences were observed between observed and expected genotype counts in either group.
The analysis of IL-23 rs2201841 gene SNP revealed no significant differences in genotypes (AA, AG, GG) or alleles (A, G) between patients with plaque psoriasis and the control group (p > 0.05). Specifically, the AG genotype was present in 32.5% of psoriasis patients and 22.5% of controls (p = 0.239), while the GG genotype was found in 12.5% of psoriasis patients and 7.5% of controls (p = 0.333). The dominant model (AG + GG vs. AA) showed no significant association (p = 0.166), and the recessive model (GG vs. AA + AG) also revealed no significant difference (p = 0.457). Additionally, the allele distribution did not differ significantly (p = 0.138), with allele A being more common in both groups (Table 2).
Table 2
IL-23 rs2201841 gene single nucleotide polymorphism among patients with plaque psoriasis and the control group
The association between the IL-23 rs2201841 gene SNP and various factors in plaque psoriasis patients was assessed. No significant associations were found between the genotypes and gender, age, affected site, duration of psoriasis, family history, or PASI score (p > 0.05 for each). However, patients with the GG genotype had the highest PASI score, while those with the AA genotype had the lowest, but this difference was not statistically significant. Additionally, a significant association was found between the GG genotype and a higher incidence of Streptococcus pyogenes (p = 0.02), while no other significant associations were observed with culture results (Table 3).
Table 3
Association between IL-23 rs2201841 gene single nucleotide polymorphism and personal history, present and family history, PASI score, and culture results in patients with plaque psoriasis
| Parameter | IL-23 rs2201841 | Test | P-value | ||
|---|---|---|---|---|---|
| AA (N = 22) | AG (N = 13) | GG (N = 5) | |||
| Gender | |||||
| Male | 13 (59.1%) | 7 (53.8%) | (3 60%) | χ2 = 0.249 | 1.000 |
| Female | 9 (40.9%) | 6 (46.2%) | (2 40%) | ||
| Age [years] | 36.41 ±9.58 | 35.08 ±11.39 | 34.60 ±5.32 | H = 1.059 | 0.589 |
| Site | |||||
| Upper limbs | 11 (50.0%) | 6 (46.2%) | 3 (60%) | χ2 = 0.385 | 1.000 |
| Lower limbs | 8 (36.4%) | 7 (53.8%) | 2 (40%) | χ2 = 1.132 | 0.667 |
| Trunk | 3 (13.6%) | 1 (7.7%) | 0 (0%) | χ2 = 0.600 | 1.000 |
| Head | 3 (13.6%) | 0 (0%) | 0 (0%) | χ2 = 1.802 | 0.530 |
| Duration of psoriasis [years] | |||||
| Median | 3.0 (0.17–10.0) | 4.0 (0.08–12.0) | 8.0 (2.0–10.0) | H = 2.598 | 0.273 |
| Family history | |||||
| No | 18 (81.8%) | 10 (76.9%) | 3 (60%) | χ2 = 1.381 | 0.577 |
| Yes | 4 (18.2%) | 3 (23.1%) | 2 (40%) | ||
| PASI score | |||||
| Median | 4.65 (0.70–19.70) | 5.30 (1.60–11.20) | 11.60 (3.0–20.70) | H = 1.776 | 0.412 |
| Any growth | |||||
| Bacterial growth alone | 7 (31.8%) | 2 (15.4%) | 1 (20%) | χ2 = 1.253 | 0.678 |
| Fungal growth alone | 5 (22.7%) | 3 (23.1%) | 0 (0%) | χ2 = 1.429 | 0.732 |
| Both bacterial and fungal growths | 10 (45.5%) | 8 (61.5%) | 4 (80%) | χ2 = 2.297 | 0.325 |
| Multiple growth | |||||
| Single | 12 (54.5%) | 5 (38.5%) | 1 (20%) | χ2 = 2.297 | 0.325 |
| Mixed | 10 (45.5%) | 8 (61.5%) | 4 (80%) | ||
| Bacterial growth | |||||
| Negative | 5 (22.7%) | 3 (23.1%) | 0 (0%) | χ2 = 1.060 | 0.730 |
| Positive | 17 (77.3%) | 10 (76.9%) | 5 (100%) | ||
| Bacterial culture | |||||
| Staphylococcus aureus | 8 (36.4%) | 8 (61.5%) | 1 (20%) | χ2 = 3.094 | 0.260 |
| Staphylococcus epidermis | 3 (13.6%) | 0 (0%) | 0 (0%) | χ2 = 1.802 | 0.530 |
| Staphylococcus saprophyticus | 1 (4.5%) | 0 (0%) | 0 (0%) | χ2 = 1.372 | 1.000 |
| Streptococcus pyogenes | 4 (18.2%) | 2 (15.4%) | 4 (80%) | χ2 = 7.484* | 0.02* |
| Corynebacterium acne | 1 (4.5%) | 0 (0%) | 0 (0%) | χ2 = 1.372 | 1.000 |
| Fungal growth | |||||
| Negative | 7 (31.8%) | 2 (15.4%) | 1 (20%) | χ2 = 1.253 | 0.679 |
| Positive | 15 (68.2%) | 11 (84.6%) | 4 (80%) | ||
| Fungal culture | |||||
| Candida albicans | 9 (40.9%) | 4 (30.8%) | 2 (40%) | χ2 = 0.374 | 0.896 |
| Aspergillus Niger | 10 (45.5%) | 8 (61.5%) | 4 (80%) | χ2 = 2.297 | 0.325 |
| Malassezia | 5 (22.7%) | 2 (15.4%) | 2 (40%) | χ2 = 1.256 | 0.590 |
IL-23 serum levels showed no significant difference between males (median = 82.89) and females (median = 57.04, p = 0.645) or associations with the affected site or family history of psoriasis. However, levels were significantly higher in patients with bacterial growth alone (median = 305.5) compared to fungal growth alone (median = 70.6) or mixed growth (median = 54.5, p < 0.001). Patients with fungal growth, including Candida albicans, Aspergillus niger, and Malassezia, had significantly lower IL-23 levels than those with negative results (p = 0.017, p < 0.001, p = 0.018, respectively). No significant differences were noted between negative and positive bacterial growth or specific bacterial cultures (Table 4).
Table 4
Association between IL-23 serum levels and gender, personal and family history, and culture results among patients with plaque psoriasis
| Parameter | IL-23 serum level | Test | P-value | ||
|---|---|---|---|---|---|
| Gender | |||||
| Male, n =23 | 175.3 ±219.1 | 82.89 | 45.72–757.0 | U = 178.0 | 0.645 |
| Female, n = 17 | 132.8 ±212.9 | 57.04 | 46.0–942.2 | ||
| Site | |||||
| Upper limbs | |||||
| No, n = 20 | 160.7 ±242.9 | 56.35 | 45.72–942.2 | U = 227.5 | 0.461 |
| Yes, n = 20 | 153.8 ±188.8 | 86.50 | 47.59–757.0 | ||
| Lower limbs | |||||
| No, n = 23 | 149.3 ±209.1 | 80.83 | 46.0–942.2 | U = 194.0 | 0.978 |
| Yes, n = 17 | 168.0 ±228.3 | 56.13 | 45.72–757.0 | ||
| Trunk | |||||
| No, n = 36 | 167.7 ±224.2 | 70.03 | 45.72–942.2 | U = 48.5 | 0.302 |
| Yes, n = 4 | 63.51 ±17.81 | 55.52 | 52.90–90.10 | ||
| Head | |||||
| No, n = 37 | 141.1 ±179.1 | 58.43 | 45.72–757.0 | U = 58.0 | 0.923 |
| Yes, n = 3 | 356.3 ±507.6 | 80.83 | 46.0–942.2 | ||
| Family history | |||||
| No, n = 31 | 178.7 ±239.2 | 80.83 | 45.72–942.2 | U = 117.5 | 0.483 |
| Yes, n = 9 | 83.38 ±49.22 | 57.04 | 47.59–174.1 | ||
| Any growth | |||||
| Bacterial growth alone | 421.25 ±307 | 305.53 | 99.59–942.15 | U = 6 | < 0.001* |
| Fungal growth alone | 79.04 ±26.37 | 70.62 | 55.65–117.32 | U = 52.9 | < 0.001* |
| Both bacterial and fungal growths | 65.67 ±27.03 | 54.54 | 45.72–144.78 | U = 44 | < 0.001* |
| Multiple growth | |||||
| Single | 269.15 ±284.25 | 130.74 | 55.65–942.15 | U = 44 | < 0.001* |
| Mixed | 65.67 ±27.03 | 54.54 | 45.72–144.78 | ||
| Bacterial growth | |||||
| Negative | 79.04 ±26.37 | 70.62 | 55.65–117.3 | U = 118.0 | 0.753 |
| Positive | 176.8 ±236.4 | 58.83 | 45.72–942.2 | ||
| Bacterial culture | |||||
| Staphylococcus aureus | |||||
| Negative | 115.9 ±119.2 | 59.22 | 47.59–584.1 | U = 178.5 | 0.645 |
| Positive | 213.1 ±295.2 | 55.43 | 45.72–942.2 | ||
| Staphylococcus epidermis | |||||
| Negative | 156.2 ±221.3 | 58.43 | 45.72–942.2 | U = 68.0 | 0.557 |
| Positive | 169.8 ±131.8 | 144.8 | 52.29–312.3 | ||
| Staphylococcus saprophyticus | |||||
| Negative | 159.8 ±216.9 | 59.22 | 45.72–942.2 | – | – |
| Positive | 55.65 | ||||
| Streptococcus pyogenes | |||||
| Negative | 180.2 ±243.1 | 70.62 | 45.72–942.2 | U = 133.0 | 0.612 |
| Positive | 88.51 ±50.33 | 58.83 | 47.59–174.1 | ||
| Corynebacterium acne | |||||
| Negative | 146.3 ±206.0 | 58.43 | 45.72–942.2 | – | – |
| Positive | 584.1 | ||||
| Fungal growth | |||||
| Negative | 421.25 ±307 | 305.53 | 99.59–942.15 | U = 6 | < 0.001* |
| Positive | 69.23 ±27.08 | 55.65 | 45.72–144.78 | ||
| Fungal culture | |||||
| Candida albicans | |||||
| Negative | 211.16 ±257.96 | 99.59 | 46–942.15 | U = 102 | 0.017* |
| Positive | 67.37 ±23.99 | 55.65 | 45.72–117.32 | ||
| Aspergillus niger | |||||
| Negative | 268.66 ±284.64 | 130.74 | 48.24–942.15 | U = 58.5 | < 0.001* |
| Positive | 66.07 ±26.91 | 55.21 | 45.72–144.78 | ||
| Malassezia | |||||
| Negative | 187.02 ±236.4 | 90.1 | 45.72–942.15 | U = 66.5 | 0.018* |
| Positive | 54.66 ±4.03 | 54.08 | 46–59.22 | ||
A strong and significant negative correlation was identified between IL-23 serum levels and the PASI score (rs = –0.866, p < 0.001), suggesting that higher IL-23 levels are associated with lower disease severity as measured by PASI. In contrast, no significant correlations were observed between IL-23 levels and the patient’s age (rs = 0.207, p = 0.199) or the duration of psoriasis (rs = –0.052, p = 0.752).
IL-23 serum levels demonstrated moderate discriminatory ability in distinguishing between plaque psoriasis and the control group, with an area under the ROC curve of 0.722 (95% CI: 0.607–0.837, p < 0.001). At a cutoff value of ≤ 117.32, IL-23 serum levels achieved 72.5% sensitivity, 65.0% specificity, 67.4% positive predictive value, 70.3% negative predictive value, and 68.75% accuracy.
The logistic regression analysis was conducted to identify predictors of plaque psoriasis susceptibility. In the univariable analysis, fungal growth showed a highly significant association with susceptibility (p < 0.001, OR = 5.010, 95% CI = 2.687–9.341). This association remained significant in the multivariable analysis, where fungal growth had an adjusted odds ratio of 3.607 (95% CI = 1.581–8.230, p = 0.002). IL-23 serum levels also demonstrated a significant inverse association in both univariable (p = 0.013, OR = 0.382, 95% CI = 0.179–0.817) and multivariable (p = 0.039, OR = 0.996, 95% CI = 0.991–0.998) analyses. Conversely, bacterial growth (p = 0.084, OR = 1.726, 95% CI = 0.929–3.208) and the IL-23 rs2201841 genotype (p = 0.166, OR = 1.498, 95% CI = 0.845–2.656) did not show significant associations with susceptibility to plaque psoriasis.
The linear regression analysis was performed to identify predictors of plaque psoriasis severity. In the univariable analysis, fungal growth was significantly associated with increased disease severity (β = 5.590, p = 0.005). This association remained significant in the multivariable model, where fungal growth had an adjusted regression coefficient of β = 3.146 (p = 0.003). Additionally, IL-23 serum levels exhibited a significant inverse relationship with disease severity, with a regression coefficient of β = –0.012 (p = 0.004) in the univariable analysis and β = –0.007 (p = 0.019) in the multivariable analysis. Other factors, including age (p = 0.798), gender (p = 0.976), disease duration (p = 0.227), bacterial growth (p = 0.144), and the IL-23 rs2201841 genotype (p = 0.824), were not significantly associated with plaque psoriasis severity in either univariable or multivariable analyses.
Discussion
Psoriasis, primarily plaque psoriasis, is a chronic inflammatory disorder driven by IL-23/IL-17A dysregulation and IL23R polymorphisms, with skin microbiota dysbiosis contributing to its pathogenesis [9, 10]. This study examines skin microbiome differences and the correlation of IL-23 and its gene SNPs with psoriasis presence and severity.
Significant differences in bacterial and fungal growth were observed between psoriasis patients and controls, with higher rates of mixed growth in the psoriasis group. These findings align with Celoria et al. [11], highlighting microbiota dysregulation, influenced by genetic and environmental factors, as a key factor in psoriasis development. Similarly, Hou et al. [12] reported greater diversity and abundance in the microbial community of psoriatic lesions compared to healthy skin. Dysbiosis, or an altered skin microbiome, has been implicated in various skin disorders, including acne [13], atopic dermatitis [14], and psoriasis [15].
Our study found Staphylococcus aureus significantly more prevalent in plaque psoriasis patients than controls, consistent with Miura et al. [16], who noted S. aureus colonization in both lesional and non-lesional psoriatic skin. Non-lesional skin may also predispose to colonization [17]. However, Maire et al. [18] reported higher S. aureus colonization in control skin and non-lesional psoriatic skin, highlighting variability across studies.
We also observed a higher incidence of Streptococcus pyogenes in psoriasis patients, aligning with De Jesús-Gil et al. [19], who linked S. pyogenes infections to plaque and guttate psoriasis exacerbation. Conversely, Patrick et al. [20] reported decreased Streptococcaceae abundance in psoriatic patients, emphasizing the complex and population-dependent role of microbes in psoriasis pathophysiology.
Our findings highlight the role of bacteria like S. aureus and S. pyogenes in psoriasis, especially in genetically predisposed individuals [5]. Skin wounds from intense itching in psoriatic lesions may enable bacterial infiltration, creating an inflammatory environment and microbiota dysbiosis [21]. Interestingly, Staphylococcus epidermidis was more prevalent in controls, supporting Nørreslet et al. [22], who suggested its loss may promote pathogen colonization, worsening inflammation.
Fungal growth was significantly more common in psoriatic patients, particularly Candida albicans, Aspergillus niger, and Malassezia, consistent with prior studies linking psoriasis to increased fungal abundance [23]. These findings align with Chadeganipour et al. [24], who identified these fungi as prevalent pathogens in psoriatic cases.
Serum IL-23 levels were significantly lower in psoriasis patients compared to controls, with moderate diagnostic utility. This contrasts with Pastor-Fernández et al. [25] and Tachibana et al. [26], who reported elevated IL-23 levels in psoriatic serum and lesions, while Piaserico et al. [27] found no significant differences, reflecting variability in IL-23 findings.
One potential biological explanation is the dynamic regulation of IL-23 levels at different disease stages. Some studies reporting elevated IL-23 levels predominantly included patients with severe or active disease, whereas our cohort comprised mostly mild-to-moderate cases, with only a small fraction having severe psoriasis (PASI > 20) [28, 29]. This suggests that IL-23 may peak in early or active disease but decline as chronic inflammation progresses, potentially due to negative feedback mechanisms or immune exhaustion limiting cytokine overproduction.
Additionally, differences in IL-23 levels may be attributable to study population characteristics, including ethnic and genetic backgrounds. Variations in IL-23 receptor polymorphisms, immune system responsiveness, and microbiome composition among different populations could contribute to discrepancies in IL-23 serum concentrations. Our study population, consisting of Egyptian patients, may exhibit genetic and environmental differences compared to previous studies.
Another confounding factor is the impact of prior treatment and immune-modulating medications, even though patients in our study had not received systemic or topical psoriasis treatments in the preceding month. Residual effects of past therapies could still influence IL-23 levels. For instance, systemic corticosteroids and biologics targeting IL-23 or IL-17 pathways may lead to prolonged suppression of IL-23 production, potentially explaining the observed lower levels.
The study also found no significant difference in genotypes and alleles of the IL-23 rs2201841 gene polymorphism between patients with plaque psoriasis and healthy controls. This observation is consistent with findings by Filiz et al. [30] and Lin et al. [31], who similarly reported no significant differences in the IL-23 rs2201841 polymorphism between psoriasis patients and controls.
The current study found a non-significant increase in the GG genotype and G allele frequencies among psoriatic patients compared to healthy subjects. This finding aligns with O’Rielly et al. [32], who reported significantly higher GG genotype and G allele frequencies in psoriatic arthritis patients. Conversely, Verbenko et al. [33] noted better therapeutic outcomes in psoriasis patients with higher G allele frequencies, suggesting potential associations with disease prognosis.
The study suggested a potential link between the GG genotype of the IL-23 rs2201841 gene polymorphism and more severe psoriasis, though the association was not statistically significant. This is consistent with Filiz et al. [30], who also found no significant relationship between the PASI score and IL-23 rs2201841 polymorphism. Notably, other studies have yet to report a direct association between this polymorphism and disease severity in psoriasis.
The present study found no significant associations between IL-23 serum levels and factors such as age, gender, family history, affected sites, or disease duration. These findings are consistent with those of Filiz et al. [30], who similarly reported no significant associations between IL-23 levels and these factors in both psoriatic patients and controls. However, Kadhum et al. [34] identified a relationship between elevated IL-23 levels and a positive family history of psoriasis.
No significant association was observed between IL-23 rs2201841 genotypes and gender. In contrast, Filiz et al. [30] reported a significant link between the rs2201841 AA genotype in males and the GA genotype in females. However, no such significant association was noted in the control group.
The study also found no significant association between IL-23 rs2201841 genotypes and age, affected site, duration of psoriasis, or family history. These findings align with those of Filiz et al. [30], who reported no significant correlation between IL-23R gene polymorphism and clinical features. Differences in ethnic background may contribute to these discrepancies as polymorphisms can vary across populations, potentially influencing the relationship between this genetic variant and disease characteristics.
The investigation into the association between IL-23 rs2201841 gene polymorphism and culture results in plaque psoriasis patients revealed no significant association between genotypes and alleles. However, a non-significant increase in the PASI score was observed among patients with the GG genotype. This finding is consistent with Filiz et al. [30], who also reported no significant association between PASI scores and this polymorphism. Similarly, Magee et al. [35] and Kamel et al. [36] noted a higher incidence of more severe psoriasis in patients carrying the GG genotype.
The study identified a higher frequency of mixed bacterial and fungal growth in psoriatic patients carrying the GG genotype of IL-23 rs2201841. Additionally, a significantly higher incidence of S. pyogenes was observed in patients with the GG genotype, followed by the AG and AA genotypes. These findings suggest that mixed infections and S. pyogenes may be associated with more severe psoriasis. This is the first study to explore the association between the IL-23 rs2201841 gene polymorphism and the microbiome in psoriasis, though the relationship between IL-23 gene polymorphisms and the microbiome has been extensively studied in other diseases.
Finally, the present study found no significant association between the IL-23 rs2201841 gene polymorphism and IL-23 serum levels in both psoriatic and control groups. This observation aligns with the findings of Filiz et al. [30], who similarly reported no significant correlation between IL-23 serum levels and gene polymorphism in either group. In contrast, Hamdy et al. [37] found that IL-23 rs2201841 gene polymorphism was associated with patients compared to the controls (p = 0.001).
In this study, we utilized culture-based methods to analyse the skin microbiome due to their practicality, cost-effectiveness, and ability to provide direct identification of viable microbial species, including antibiotic susceptibility testing. However, we acknowledge the limitations of culture methods as they may not capture the full microbial diversity, particularly unculturable or fastidious organisms. Advanced techniques such as 16S rRNA sequencing and metagenomic approaches offer a more comprehensive and unbiased characterization of microbial communities by identifying both culturable and non-culturable species. These methods can also provide insights into microbial functional pathways and host-microbiome interactions, which are crucial in psoriasis pathogenesis. While 16S rRNA sequencing and metagenomics were not feasible in this study due to resource constraints, future studies integrating these advanced molecular techniques will help elucidate the broader microbial landscape in psoriasis and its potential implications in disease severity and treatment response.
In this study, the IL-23 rs2201841 polymorphism did not show a significant association with psoriasis susceptibility or severity. While this SNP has been investigated in various autoimmune conditions, including psoriasis, its role remains inconsistent across different populations. Previous studies have identified other IL-23-related polymorphisms, such as rs11209026 and rs10889677, as more strongly associated with psoriasis and psoriatic arthritis [38]. The rs11209026 (Arg381Gln) polymorphism, for instance, has been linked to impaired IL-23 receptor signalling, which could influence Th17-mediated inflammation [39]. Similarly, rs10889677, located in the IL-23R promoter region, has been associated with increased IL-23R expression and susceptibility to psoriasis. Additionally, genetic variations in other cytokine-related genes, including IL-17A (rs2275913), TNF-α (rs1800629), and IL-6 (rs1800795), have been implicated in psoriasis pathogenesis [40]. Given these findings, future studies should explore a broader range of SNPs within the IL-23/Th17 pathway and assess their combined effects on disease susceptibility, severity, and treatment response.
While this study provides valuable insights into the association between the skin microbiome, IL-23 levels, and psoriasis, several limitations must be acknowledged. First, the reliance on self-reported family history of psoriasis introduces a potential recall bias as participants may not accurately recall or report familial cases, leading to underestimation or overestimation of genetic predisposition. Second, geographic and environmental influences could impact the microbiome composition and immune responses as variations in climate, dietary habits, and healthcare access may contribute to differences in psoriasis manifestations across populations. Third, the study’s single-centre design may limit the generalizability of findings as participants were recruited from a specific geographic region with potentially distinct genetic and environmental backgrounds. Additionally, while we excluded participants who recently used systemic or topical treatments, we cannot entirely rule out the long-term effects of past treatments on microbiome composition and cytokine levels. Future studies with larger, multi-centre cohorts and more comprehensive genetic and microbiome analyses would help validate these findings and minimize potential biases.