Introduction
Gallbladder cancer (GBC) is a highly aggressive disease with marked geographical variations [1]. Northern India is amongst the regions with the highest incidence of GBC [1]. Due to the lack of symptoms, the disease is usually diagnosed at an unresectable stage. Accurate diagnosis of GBC is challenging for radiologists, especially in polypoidal and mural thickening types of lesions [2]. The mural thickening type of GBC is particularly challenging to diagnose as gallbladder wall thickening is a nonspecific finding. Recently, a Gallbladder Reporting and Data System (GB-RADS) was proposed for risk stratification of gallbladder wall thickening on ultrasound (US). This system identifies five categories (GB-RADS 1-5) with increasing risk of malignancy [3]. An additional category GB-RADS 0 includes non-diagnostic US for several reasons, including gallbladder-related factors (e.g., contracted gallbladder, gallbladder filled with calculi, gas) and patient-related factors (e.g., obesity, abdominal wounds, abdominal scar) [3]. Accurate diagnosis in this subset of patients may be significantly delayed.
Recently, a deep learning-based approach has been advocated for imaging-based diagnosis [4-7]. A few studies have shown promising results in diagnosing gallbladder cancer with US-based deep learning models [8-12]. However, to our knowledge, no study has explored the potential of deep learning in non-diagnostic US examinations. As non-diagnostic US examinations of gallbladder are common, the role of deep learning in this context must be explored. A deep learning approach may lead to early diagnosis of GBC and development of cost-effective strategies. Thus, we aimed to evaluate the diagnostic performance of a deep learning-based classification of gallbladder lesions on US images in patients with non-diagnostic US secondary to gallbladder factors (GB-RADS 0 lesions).
Material and methods
Patients
This study comprised consecutive patients with suspected gallbladder lesions who underwent gallbladder US scans as part of a prospective study on artificial intelligence (AI) based classification of GB lesions on US, which was approved by the institutional ethics committee (IEC-11/2019-1403), and all recruited patients gave informed written consent. Consecutive patients with non-diagnostic US due to calculi within the gallbladder lumen (obscuring the detailed visualization), who had the final pathological diagnosis, were included. The final diagnosis was based on fine-needle aspiration cytology, percutaneous or endoscopic biopsy, or surgical histopathology.
Patients whose final diagnosis was unavailable and those in whom the non-diagnostic US was due to patient factors were excluded. Demographic details, including age, gender, presence of biliary involvement, ascites, liver, and omental metastasis, were recorded.
Ultrasound of the gallbladder
Ultasound images were acquired by radiologists with 1-3 years of post-training experience in abdominal US. All patients were instructed to remain fasting for at least six hours before examination. US images were acquired using a convex transducer (1-5 MHz) in different planes to evaluate all gallbladder parts.
Identification of cases for inclusion in the study
A research fellow (RS) with 1 year of experience in handling gallbladder-related data (in the AI gallbladder US project mentioned above) identified the consecutive non-diagnostic US reports of gallbladder lesions. These reports were then evaluated by a radiologist (PG) with ten years of post-training experience in abdominal US who identified patients with non-diagnostic US due to calculi within the gallbladder lumen, obscuring the detailed evaluation. The radiologist (PG) was blinded to the final diagnosis. The research fellows then retrieved the images of these patients. The radiologist (PG) re-read the images to confirm the non-diagnostic nature of the US images.
Deep learning (DL) models
We used convolution neural networks (ResNet50, GBCNet), transformer models (vision transformer (ViT), RadFormer), and a hybrid model (MedViT). Of these, GBCNet (https://paperswithcode.com/paper/radformer-transformers-with-global-local) and RadFormer (https://paperswithcode.com/paper/radformer-transformers-with-global-local) have already been trained on a public dataset (https://paperswithcode.com/dataset/gbcu). We trained ResNet 50, ViT, and MedViT on a public gallbladder dataset (GBCU dataset). The GBCU dataset comprises 1255 US images from 218 patients (432 normal, 558 benign, and 265 malignant US images). For training the models, we labelled normal and benign images as class 0 (benign) and malignant images as class 1 (malignant). GBCNet and RadFormer give three classification levels (normal, benign, and malignant). We changed the classification layer to two-level prediction (benign and malignant). The testing was done on our dataset (304 images). We changed the image-level predictions to patient-level predictions using majority-level prediction [13]. All implementation was done in the PyTorch framework.
Statistical analysis
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and diagnostic accuracy of the DL models for detecting GBC were calculated. The area under the curve (AUC) for detecting GBC was plotted on the receiver operating characteristic (ROC) curve. The statistical analysis was done using IBM SPSS software.
Results
Patients
We identified 41 US reports with non-diagnostic gallbladder US. Of these, 32 non-diagnostic US reports due to gallbladder-related factors were identified. The review by the radiologist (PG) identified 29 true non-diagnostic US. Three patients without a pathological diagnosis were excluded. Thus, 26 patients (mean age [SD]: 57.5 ±8.07 years, 17 female) were included in this study. Twelve patients had benign gallbladder lesions and 14 had GBC. The total number of US images was 304.
Fifteen (57.6%) patients had abdominal pain and 8 (30.8%) patients had jaundice at the time of US (choledocholithiasis in 4). Four patients with GBC had biliary involvement, 3 had liver metastasis, and 2 had omental metastases. Ascites was present in 2 patients (Table 1). None of the patients with GBC was receiving chemotherapy at the time of US.
Table 1
Patient characteristics
Deep learning model performance
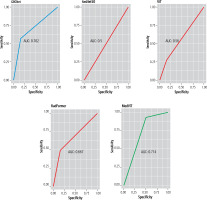
GBCNet and MedViT had the best diagnostic performance for detecting GBC and benign gallbladder lesions (Figs. 1-4). The sensitivity, specificity, and diagnostic accuracy of GBCNet for detecting GBC were 51.1% (95% confidence interval [CI]: 28.8-82.3%), 83.3% (95% CI: 51.5-97.9%), and 70% (95% CI: 48.2-85.6%), respectively. The PPV and NPV were 80% (95% CI: 51-93.8%) and 62.5% (95% CI: 46.3-76.2%), respectively. The AUC was 0.709 (95% CI: 0.541-0.873) (Fig. 3). The sensitivity, specificity, diagnostic accuracy, PPV, NPV, and AUC of MedViT for detecting GBC were 92.8% (95% CI: 66.1-99.8%), 50% (95% CI: 21-78.9%), 73% (95% CI: 54.7-79.5%), 68.4% (95% CI: 54.7-79.5%) and 85.7% (95% CI: 45.5-97.7%), respectively. The AUC was 0.714 (95% CI: 0.550-0.877). For benign thickening, GBCNet had high sensitivity (83.3%) and MedViT had high specificity (92.8%). There was no significant difference between the performance of GBCNet and MedViT. The other models had inferior diagnostic performance compared with GBCNet and MedViT for detecting GBC (Table 2).
Table 2
Diagnostic performance of deep learning models for detecting gallbladder cancer in test cohort
Fig. 1
Accurate diagnosis of gallbladder cancer with GBCNet. A) Ultrasound image of a 55-year-old man who presented with jaundice shows a wall-echo-shadow complex. The ultrasound was reported as non-diagnostic. The patient underwent contrast-enhanced MRI. B) T2-weighted MRI shows the gallbladder lumen filled with calculi (arrows). C) Diffusion-weighted image at b = 800 shows mild diffusion restriction within the gallbladder wall. D) Contrast-enhanced image shows asymmetric enhancing mural thickening. The patient underwent a biopsy of an omental nodule (not shown) that was reported as metastatic adenocarcinoma. This case was correctly predicted as cancer by GBCNet while MedViT predicted it as benign (false negative)
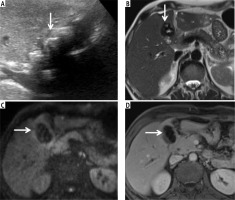
Fig. 2
Accurate diagnosis of benign gallbladder lesion with MedViT. Ultrasound image wall-echo-shadow complex (arrow, A). Axial T2-weighted magnetic resonance image (MRI) shows large calculi within the gallbladder lumen (arrows, B). Axial (C) and coronal (D) contrast enhanced MRI show thin smooth mural enhancement (arrows). Cholecystectomy revealed changes of chronic cholecystitis. GBCNet predicted this case as malignant (false positive)
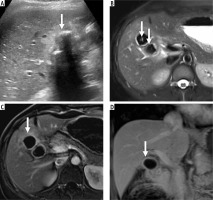
Fig. 3
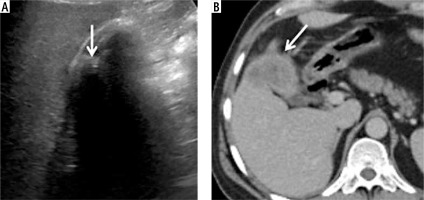
Accurate diagnosis of gallbladder cancer with MedViT. Ultrasound image is nondiagnostic due to the shadowing by the calculi (arrow, A). Patient underwent non-contrast computed tomography (CT) as she had deranged renal function. Axial (B) and coronal reformatted (C) CT image shows marked asymmetric mural thickening of the gallbladder. D) Axial CT image at a caudal level shows omental nodules. Omental biopsy revealed metastatic adenocarcinoma. GBCNet predicted this case as benign (false negative)
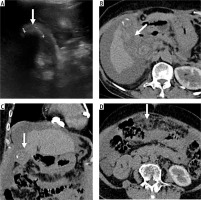
Fig. 4
False negative diagnosis with GBCNet and MedViT. The ultrasound image shows the wall-echo-shadow complex (arrow, A). The patient underwent a contrast-enhanced CT scan that showed asymmetrical mural thickening with infiltration of adjacent liver parenchyma (arrow, B). Extended cholecystectomy revealed gallbladder adenocarcinoma not otherwise specified. In this case, both GBCNet and MedViT gave benign predictions
Discussion
In this study evaluating the performance of the deep learning models in classifying gallbladder lesions in patients with non-diagnostic US, we found that GBCNet had high specificity and PPV and MedViT had high sensitivity and NPV for detecting GBC. These results suggest that the deep learning models can potentially stratify patients with non-diagnostic US. However, further improvement in the performance is needed to render this approach relevant in clinical practice.
Ultrasound is the first-line imaging modality for evaluating patients with suspected gallbladder lesions [14]. US is a widely available and relatively inexpensive test. In patients with normal gallbladder in US, no further imaging evaluation is needed. Similarly, patients with benign diagnoses on US can be followed up clinically. Patients with equivocal and malignant lesions on US undergo contrast-enhanced CT or MRI [3]. Non-diagnostic US can be due to several reasons, including gallbladder and patient-related factors. Subjecting all these patients to contrast enhanced CT or MRI is not cost-effective as most patients with non-diagnostic US have benign gallbladder lesions e.g., chronic cholecystitis [15]. The risk stratification of this group of patients is highly desirable.
Deep learning-based approaches can potentially improve radiologists’ diagnostic performance. Deep learning has shown promising results in evaluating gallbladder lesions on US [8-12]. A recent study compared the performance of a CNN-based model for classifying gallbladder polyps into neoplastic and non-neoplastic with three radiologists on US. The CNN showed a sensitivity, specificity, accuracy, and AUC of 74.3%, 92.1%, 85.7%, and 0.92 for neoplastic polyps [8]. The AUC of CNN for detecting neoplastic polyps was comparable to the radiologists (0.78-0.94). In another study utilizing CNN for classifying gallbladder polyps as neoplastic and non-neoplastic on endoscopic US, the sensitivity and specificity of CNN were 60.3% and 77.4%, respectively, compared to 74.2% and 44.9%, for the endoscopists [9]. A state-of-the-art CNN model based on multiscale, second-order pooling architecture (GBCNet) was recently proposed [10]. The sensitivity, specificity, and accuracy of GBCNet to classify images as normal, benign, and malignant were 92.9%, 90%, and 87.7%, respectively. The accuracy of GBCNet for binary classification of images into malignant and non-malignant was substantially higher than that of the two radiologists (91% vs. 78.4-81.6%). Another study reported accuracy of the DL-model comparable to expert radiologists in detection of GBC [12]. The DL model performed well even on extensive subgroup analysis [12]. However, to our knowledge, the performance of deep learning models in gallbladder lesion classification in patients with non-diagnostic US has not been investigated.
There were a few limitations to our study. First, the sample size was small. However, we included only those patients who had a pathological diagnosis. Second, not all patients underwent cholecystectomy. Thus, fine needle aspiration cytology and core biopsy results were used as the reference standard in addition to surgical histopathology. Third, we could not evaluate the explainability of all the DL models. RadFormer has been previously reported to focus on important areas mentioned in GB-RADS, but for other models, such work should be done in future. Finally, we did not test the performance of these models on an external dataset.
In conclusion, the deep learning approach has the potential to improve gallbladder lesion classification in patients with non-diagnostic US. However, the performance of deep learning models needs to be enhanced for them to be incorporated into clinical practice. Prospective multicentre studies using histopathology as the reference standard may help achieve this.