Introduction
Psoriasis is a common, chronic immune-mediated skin disease. Clinically, psoriasis is characterized by the presence of well-demarcated, scaly, erythematous skin lesions, and is frequently associated with arthritis [1]. Psoriatic arthritis is a seronegative inflammatory arthritis present in nearly 25% of patients with psoriasis and develops after an average interval of about one decade [2]. Psoriatic arthritis is defined as a subtype of spondyloarthropathies, based on common human leukocyte antigen (HLA) associations. Clinically, this disease is characterized by changing degrees of oligoarthritis, polyarthritis and spondylitis and typically involves dactylitis, distal interphalangeal joint involvement or mutilating arthritis [2, 3].
Early onset psoriasis, or type I psoriasis, refers to patients with an onset before 40 years of age, more serious disease course than the late onset psoriasis, strong family history, and HLA-C*06 positivity. Early onset psoriasis has been reported in 75% of patients with psoriasis; hence, it affects the majority of patients with psoriasis. Late onset psoriasis, or type II psoriasis, in contrast, is characterized by an onset at or after 40 years, less severe clinical symptoms and rare family inheritance [4, 5]. Recently, age at disease onset (with a cut-off value at 40 years) has been found similarly important in the characterization of the disease phenotype in psoriatic arthritis as well [6].
Single nucleotide polymorphisms (SNPs) of several genes have been identified as contributing to psoriasis susceptibility [7, 8]. We and others have previously reported a significant association with variants of the HLA-Cw*0602, IL23R, LCE3C, LCE3B-del [9, 10], IL12R, IL23 [11], TNFSF15 [3], and ERAP1 genes [12–14]. Psoriatic arthritis is strongly associated with the HLA-B13, HLA-B57, HLA-B39, HLA-Cw6 and HLA-Cw7 alleles [8]. While psoriatic arthritis tends to have a more severe disease course and appears earlier in HLA-B*27 positive patients, the latency between the onset of psoriasis and onset of joint symptoms is longer in HLA-Cw*0602 positive patients [6, 15].
Genetic variants of the endoplasmic reticulum-associated amino-peptidase 1 (ERAP1) gene has recently been reported to be associated with ankylosing spondylitis (AS), psoriasis [16], and Behcet’s disease [17]. The ERAP1 protein belongs to the M1 family of zinc metallopeptidase enzymes and is encoded by a gene on chromosome 5q15 [18]. ERAP1 trims peptides imported into the endoplasmic reticulum at their N-terminus and contributes to the shaping of the antigenic repertoire presented by class I major histocompatibility complex (MHC) molecules [19]. Depending on the peptide length and sequence composition, ERAP1 has the ability to both destroy and create peptide cargos for MHC class I molecules [20]. The ERAP1 protein also contributes to the shedding of the membrane-bound receptors of inflammatory cytokines, such as IL-1R2, TNFR1 and IL-6R [21]. ERAP1 likely plays a pivotal role in the protection from infectious diseases by contributing to the maintenance of immune tolerance and control of inflammation [22]. The association of ERAP1 variants with psoriasis has been investigated recently in populations of European and Chinese ancestry. Two ERAP1 variants, rs27524 (noncoding) and rs30187 (Lys528ARG), were found to be genome-wide risk factors for psoriasis [12]. Furthermore, dominant epistasis between HLA-Cw*0602 and the ERAP1 rs30187 SNP was identified in studies [12–14]. Another recent work identified the ERAP1 rs27432 SNP (intronic) as a variant strongly associated with psoriasis [23]. In Han Chinese populations, ERAP1 SNPs or gene variants in linkage disequilibrium with ERAP1 SNPs were also found to be associated with psoriasis [24–26]. It is currently unclear whether the effects of ERAP1 on psoriasis can be explained by a single variant or by allotypic associations. Recently, the association of ERAP1 with psoriatic arthritis has also been investigated; however, this study did not demonstrate an association between rs30187 and psoriatic arthritis or psoriatic arthritis subphenotypes [27].
As psoriasis is currently best stratified by disease onset and absence or presence of arthritis, we hypothesized that stratifying psoriasis patients into early and late onset groups, as well as skin-only and arthritis subgroups leads to the identification of new ERAP1 genotype–phenotype associations. Furthermore, as ERAP1 seems to be associated with inflammatory diseases in a HLA-dependent manner, we also intended to explore whether a gene–gene interaction between HLA-Cw*0602 and ERAP1 exists in these well-stratified subgroups of psoriatic patients.
Aim
Furthermore, as ERAP1 seems to be associated with inflammatory diseases in a HLA-dependent manner, we also intended to explore whether a gene–gene interaction between HLA-Cw*0602 and ERAP1 exists in these well-stratified subgroups of psoriatic patients. We have analysed the association of five ERAP1 and two HLA-Cw*0602 SNPs as well as different ERAP1 haplotypes with EOP, LOP and psoriatic arthritis.
Material and methods
Subjects
This study was approved by the Internal Review Board of the University of Szeged, the approval number is PSO-GENET-001. Informed consent was obtained from all participating patients and volunteers, and the study was conducted in full accordance with the principles of the Declaration of Helsinki. The study population consisted of 319 Hungarian Caucasian psoriasis vulgaris patients (designated as PsV) treated at the Department of Dermatology and Allergology and the Department of Rheumatology of the University of Szeged, and at the Department of Rheumatology of Pándy Kálmán Békés County Hospital, Gyula, as well as 200 ethnically-matched healthy individuals with no known multifactorial inflammatory diseases. Of the 319 psoriasis vulgaris patients, 105 exhibited psoriatic arthritis (designated as PsA), fulfilling the Classification Criteria for Psoriatic Arthritis (CASPAR). The group of patients with psoriatic arthritis was further stratified into five homogenous clinical groups according to the Moll and Wright criteria [28]. Patients with skin symptoms of psoriasis only were denoted as cutaneous psoriasis (designated as PsC) patients. Patients with onset before 40 years of age were classified as having early onset psoriasis (EO-PsV, EO-PsA, EO-PsC), whereas late onset psoriasis (LO-PsV, LO-PsA, LO-PsC) was defined by an onset at or after 40 years. The demographic and clinical characteristics of the study population are listed in Table 1.
Table 1
Demographic and clinical characteristics of the study population
Genotyping and haplotype analysis
Genomic DNA was isolated from venous blood of patients and controls using the BioRobot EZ and the EZ1 DNA Blood Kit from QIAGEN (Hilden, Germany), according to the instructions of the manufacturer. Genotyping of five SNPs of the ERAP1 gene and two SNPs of the HLA-Cw*0602 gene (Table 2), previously reported as candidate genes and SNPs in the pathogenesis of psoriasis and AS [16, 29], was carried out with the PCR-based Assay-by-Design method of Applied Biosystems (Foster City, CA), following the instructions of the manufacturer. After PCR amplification, end-point detection was performed with a CFX 96 real-time PCR machine from Bio-Rad (Hercules, CA). Genotyping of the rs10484554 SNP was used to determine the HLA-C status, as previously reported [12, 16]. Three SNPs (rs30187, rs10050860, rs17482078) were used to construct four haplotypes: Haplotype A (rs17482078/rs10050860/rs30187-CCC), Haplotype B (rs17482078/rs10050860/rs30187-CCT), Haplotype C (rs17482078/rs10050860/rs30187-TTC) and Haplotype D (rs17482078/rs10050860/rs30187-TTC, previously reported by Ombrello et al. to be risk or protective factors in AS [30].
Table 2
ERAP1 and HLA-Cw*0602 gene SNPs
Statistical analysis
Genotype frequencies, frequencies of the main haplotypes and SNP associations were calculated and compared between patient groups using Fisher’s exact test, using the Plink software package (v1.9). Odds ratios were calculated with 95% confidence intervals. Multiple-testing correction was applied to all comparisons using the Benjamini-Hochberg ‘FDR’ method in R (v3.2.3), with a significance threshold of p < 0.05. As previously suggested and successfully applied by other authors [31–33], the age of 40 years was used for stratification of age at disease onset. No further stratification of age at disease onset was attempted due to the relatively small number of patients.
Results
The genotype frequencies of the HLA-C and ERAP1 SNPs are summarized in Table 3. Genotype frequencies in patients and controls were in Hardy-Weinberg equilibrium. Control genotype frequencies were comparable to those published in the literature. The genotype distribution of one HLA-C SNP (rs10484554) was found to be significantly different between the PsV patients and the group of healthy individuals (p = 5.9 × 10–5 respectively). The proportion carrying the mutant HLA-Cw*0602 allele (rs10484554 SNP) was significantly higher among PsV patients than in the group of healthy controls (58.3% and 36.5%, respectively), and there was no difference in this respect between patients with skin only or skin and joint symptoms (57.9% and 59%, respectively). Taken together, the HLA-Cw*0602 rs10484554 SNP seems to be a strong susceptibility factor for psoriasis (Table 3). In the case of the five other ERAP1 SNPs (rs27524, rs27525, rs30187, rs17482078 and rs10050860) and rs10484545 HLA-C SNP, there were no statistically detectable differences in the genotype distributions between healthy individuals and the PsV patients.
Table 3
Distribution of ERAP1 and HLA-Cw SNP genotypes in healthy controls and psoriasis patients
Subsequently, psoriasis vulgaris patients were subdivided into groups according to the presence or absence of arthritis (Table 3). In line with earlier publications [29, 32, 34], the genotype distribution of the HLA-C rs10484554 SNP was found to be significantly different in both the PsC and the PsA subpopulations (p = 0.0007 and p = 0.0007, respectively), compared to healthy controls. In PsA (but not in PsC) patients, the genotype distribution of three ERAP1 SNPs (rs10050860, rs27525 and rs17482078) was also significantly different (p = 0.0252, p = 0.0453 and p = 0.0453, respectively) (Table 3). For all three SNPs, the proportion of patients carrying the wild type allele was higher than in the group of healthy individuals, suggesting that the rare allele of these SNPs might provide protection against the development of psoriatic arthritis. As no differences were detected in the genotype distribution of the studied ERAP1 SNPs in PsC patients, it is likely that the trend in the difference observed for the rs10050860 SNP in the PsV patients is caused by the presence of the PsA patients. No difference was detected for the ERAP1 rs27524 and rs30187 SNPs and the HLA-C rs10484545 SNP between the healthy controls and the psoriasis groups, even after stratification for arthritis.
We further stratified patients according to disease onset (early and late onset). Significant differences were observed only for the HLA-C rs10484554 SNP in the early onset groups, which is in agreement with earlier publications [13, 14, 29, 34]. This result suggests that HLA-C positivity is a susceptibility factor only for early onset psoriasis. Stratification led to no significant difference for the rs10484545 HLA-C SNP. The ERAP1 SNPs (rs10050860, rs27525 and rs17482078) known to be associated with psoriatic arthritis were found to have only a tendency in the association with EO-PsA patients (p = 0.0663, p = 0.0663 and p = 0.0997, respectively) (Table 3). This lends further support to the notion that the differences in distribution of these SNPs detected in the total PsA group are primarily driven by the early onset subpopulation, and that these ERAP1 SNPs presumably provide protection against early onset psoriatic arthritis.
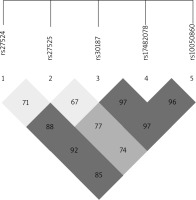
The ERAP1 rs17482078, rs10050860, rs30187 and rs2287987 SNPs were found to be in strong linkage disequilibrium, and the association between haplotypes including these SNPs and AS was recently reported [35]. A linkage disequilibrium block containing the rs17482078/rs10050860/rs30187 SNPs was also identified in our dataset (Figure 1). Thus, we examined whether the rs17482078/rs10050860/rs30187 haplotypes were associated with psoriasis susceptibility (Table 4). We found that Haplotype B (rs17482078/rs10050860/rs30187-CCT) was a risk factor only for LO-PsV (p = 0.0409) and for LO-PsA (p = 0.0413).
Table 4
Distribution of ERAP1 haplotypes in healthy controls and psoriasis patients
As it was reported earlier that some ERAP1 variants influence psoriasis susceptibility exclusively in individuals carrying the HLA-C risk allele [12–14], we analysed ERAP1 SNPs in HLA-C positive psoriasis and psoriatic arthritis patients. ERAP1 SNP frequencies were compared between individuals carrying at least one copy of the risk allele of rs10484554 (HLA-C positive) and individuals not carrying the HLA-C sequence (HLA-C negative) (Table 5). Evidence for association in HLA-C positive individuals was observed for two loci. The ERAP1 rs27524 SNP in HLA-C positive individuals exhibited a 1.74-fold increased risk of PsV (p = 0.0454) and a 2.33-fold risk of PsA (p = 0.0185), in agreement with the previous report [12]. Interestingly, rs27524 by itself was not associated in our dataset with either PsV or PsC and was found to have a tendency as a susceptibility factor only for LO-PsA (Table 3). The ERAP1 rs27525 SNP decreased the risk of psoriatic arthritis development in HLA-C positive patients (odds ratio (OR) 0.42, p = 0.0339). Thus, the presence of the ERAP1 rs27525 SNP seems to protect HLA-C positive individuals from developing psoriatic arthritis. No other interaction was found for the ERAP1 rs30187, rs10050860 and rs17482078 SNPs in HLA-C positive individuals, even after further stratifying the patients into early and late onset subgroups.
Table 5
HLA-C and ERAP1 interactions in psoriasis and psoriatic arthritis
* Individuals carrying at least one copy of the rare allele of the rs10484554 HLA-C SNP and both wild type alleles of the indicated ERAP1 SNP (HLA-C positive/ERAP1 negative) are compared with individuals carrying at least one copy of the rare allele of the rs10484554 HLA-C SNP and at least one copy of the rare allele of the indicated ERAP1 SNP (HLA-C positive/ERAP1 positive).
Discussion
Psoriasis has been associated with the PSORS1 disease susceptibility locus of the MHC class I region on chromosome 6. Within PSORS1, the HLA-Cw6 gene has shown the strongest association with psoriasis [8]: about 60% of psoriasis patients carry the HLA-Cw*0602 allele. HLA-C positive patients exhibit earlier disease onset, higher incidence of guttate and eruptive type of psoriasis, more extensive disease symptoms, and more frequent exacerbations caused by throat infections than HLA-C negative individuals. The frequency of HLA-Cw*0602 is significantly lower in patients with psoriatic arthritis compared with those with psoriasis alone, and its presence is associated with a longer psoriasis–arthritis interval and milder arthritis forms [34, 36, 37].
In this study, a well-characterized cohort of psoriasis patients was available for investigating whether HLA-Cw6 or ERAP1 gene polymorphisms are associated with different clinical phenotypes of psoriasis and psoriatic arthritis. Our dataset included patients with early and late onset psoriasis vulgaris, as well as with cutaneous psoriasis and psoriatic arthritis.
Our results confirmed the previously reported genetic association of psoriasis with both HLA-Cw6 and ERAP1 genes [12–14]. The HLA-Cw*0602 rs10484554 SNP was found to have a very strong association with PsV, and the association was highly significant with both PsC and PsA groups as well (although somewhat stronger with PsC). Interestingly, only EO-PsV was associated with HLA-C in our dataset. Previously, the HLA-Cw*06 allele was not found to be a risk factor for late onset psoriasis in a Northern Polish population [38], and late onset psoriasis demonstrated only a weak association with HLA-Cw*06 alleles in other studies [13, 39]. More recently, however, a study using dense genotyping revealed that HLA-Cw*06 is associated with late onset psoriasis [40].
Genetic variants of ERAP1 have recently been reported to be associated with psoriasis [13, 14, 16]. ERAP1 trims peptides imported into the endoplasmic reticulum at their N-terminus and contributes to the shaping of the antigenic repertoire presented by class I MHC molecules [19]. The association of ERAP1 with psoriatic arthritis has previously been investigated; however, no association between ERAP1 SNPs and psoriatic arthritis or psoriatic arthritis subphenotypes was identified [27]. Recently, ERAP1 association with psoriasis was detected in patients with an age of onset between 10 and 20 years, and no association was detected in cases with onset below 10 years [16].
Our results suggest that the HLA-C rs10484554 SNP contributes to psoriasis susceptibility. The proportions of the rare alleles of the ERAP1 rs27525, rs17482078 and rs10050860 SNPs were higher in the group of healthy individuals, suggesting that individuals with the rare alleles of these SNPs might be protected against psoriasis. These associations were also apparent when groups of psoriatic patients were analysed separately according to the presence or absence of arthritis. The stratification of the patients for early (< 40 years) or late (≥ 40 years) disease onset revealed an age-dependent difference in the genetic background of psoriasis: the associations with these SNPs tend to be stronger in patients with early disease onset.
Although ERAP1 was neither dependent on nor interacting with HLA-C*06:02 in certain populations, an interaction between HLA-C and ERAP1 was reported recently and confirmed several times subsequently [9, 12–14]. Interestingly, dominant epistasis between HLA-Cw*0602 and one of the ERAP1 SNPs was identified recently in psoriasis [23].
Recent studies of the ERAP1 rs27524 SNP reported association with the pathogenesis of psoriasis, especially in the group with age of disease onset between 10 and 20 years [16, 41]. Our results revealed only a trend in association with the late disease onset group, for which it might increase the likelihood of psoriasis development. No association with psoriasis was observed for the ERAP1 rs30187 SNP in this study, which is in contrast with previous studies [12, 16]; however, Jadon et al. also reported that there was no association with psoriatic arthritis [27]. In addition, this psoriasis-risk allele, which is common in the European population, was not associated with the disease in an East Asian population [24].
Psoriatic arthritis patients, especially in the early onset group, carrying the ERAP1 rs27525 and rs17482078 SNPs seem to be protected from the subsequent development of the disease. Individuals carrying the rare allele of the ERAP1 rs10050860 SNPs might be protected against psoriasis, but this effect was more prominent among patients with psoriatic arthritis, especially in the early onset group. These findings suggest that these three SNPs (rs27525, rs17482078, rs10050860) might be protective against psoriatic arthritis.
The two coding ERAP1 SNPs (rs17482078 and rs10050860) and the rs30187 SNP are in linkage disequilibrium (Figure 1) and have been reported as a protective haplotype (rs17482078/rs100508607rs30187/rs2287987–TTCC) in HLA-B positive AS patients [35]. Also the rs17482078/rs10050860/rs30187-CCT haplotype was confirmed as a risk factor for AS [42] in an AS population in Belgium, which we examined as Haplotype B (rs17482078/rs10050860/rs30187-CCT) in psoriasis susceptibility similarly to AS [35], and it was found to be a risk factor for only LO-PsV (p = 0.0409) and LO-PsA (p = 0.0413). We were unable to find any association with psoriasis or with psoriatic arthritis for Haplotype A (rs17482078/rs10050860/rs30187-CCC), Haplotype C (rs17482078/rs10050860/rs30187-TTC) and Haplotype D (rs17482078/rs10050860/rs30187-TTC). These data suggest that the Haplotype B conferring disease risk in AS also influences susceptibility to joint involvement in psoriasis.
Genome-wide association studies identified the interaction between the ERAP1 rs27524 SNP and the HLA-C rs10484554 SNP; this interaction was the most prominent among individuals carrying one or two copies of the risk allele at rs10484554 [12]. In subsequent analysis, the association with ERAP1 was not restricted to individuals carrying HLA-Cw*0602; however the genetic association with ERAP1 (rs207524, rs30187, rs26653) in psoriasis was confined to individuals with an age of disease onset between 10 and 20 years [16]. In a Polish population, ERAP1 rs27524 SNP was a susceptibility factor for HLA-C positive patients with late onset psoriasis [13]. In our study, the ERAP1 rs27524 SNP in HLA-C positive individuals caused a 1.74-fold increased risk of PsV (p = 0.0454) and a 2.33-fold risk of PsA (p = 0.0185). Notably, the rs27524 SNP by itself was not associated either with PsV or with PsC in this dataset and was found to have only a tendency as a susceptibility factor for LO-PsA (Table 3). The ERAP1 rs27525 SNP in HLA-C positive patients decreased the risk of psoriatic arthritis development (OR = 0.42, p = 0.0339); thus, the presence of these SNPs seems to protect against developing psoriatic arthritis.
Taken together, our results suggest that the genetic variants of the ERAP1 and HLA-C genes contribute to the pathogenesis of psoriasis in a manner that is dependent on age of onset. Individuals with HLA-Cw*0602 are more prone to early onset of disease (before 40 years), confirming that onset after 40 years represents a biologically valid approximation for a genetically distinct subgroup of psoriasis. The overall psoriasis group was stratified by various clinical aspects, including the age of onset and the presence or absence of psoriatic arthritis. This novel and careful stratification of patients according to the symptoms and age of onset leads to important insights for psoriasis, a heterogeneous, multifactorial disease, and might become more important for further research as well as for personalized medicine.