Introduction
Hepatocellular carcinoma (HCC) is a global health issue and is the most common type of liver cancer, accounting for 90% of all cases [1]. In 2020, HCC was estimated to be the third leading cause of cancer death worldwide, with only a slight difference between incidence rate and mortality rate, reflecting the poor prognosis of HCC [2, 3]. In Egypt, it represents the fourth most common cancer and the most common mortalityand morbidity-related cancer [4].
Hepatocellular carcinoma develops in most cases on top of cirrhosis except in the case of hepatitis B virus (HBV) which has the unique ability to develop HCC without cirrhosis [5]; thus the prognosis in HCC depends mainly on the underlying liver cirrhosis, which limits the available options of treatment as it may affect the remaining normal liver tissue [6-8]. Hence, surveillance for HCC among high-risk groups, mainly patients with established cirrhosis, is crucial [8-10]. Ultrasound assessment is used for this aim with satisfactory pooled sensitivity, though it still has its limitations; the most important limitation is liver cirrhosis itself, which disrupts the liver architecture, making the detection of small focal lesions more challenging and requiring much experience [8, 11].
Alpha-fetoprotein (AFP) is the gold standard biomarker in HCC, but its levels are affected by flares of HBV and hepatitis C virus (HCV) or deterioration of liver condition in addition to its low sensitivity in detection of early and small HCC [12]. The addition of AFP to ultrasound was found to significantly increase false positive results [11, 13]. As a result, most of the guidelines do not recommend the use of AFP with or without ultrasound in surveillance for HCC on regular bases and the identification of other biomarkers is needed, especially for the diagnosis of early HCC [9, 10].
Long non-coding RNAs (lncRNAs), defined as noncoding RNA longer than 200 nucleotides [14], act as regulators in many physiological processes, such as the regulation of cell proliferation, differentiation and apoptosis [15]. Recently it was found that they play a critical role in oncogenesis, tumor suppression and invasion of different types of cancers, of which HCC was not an exception [16]. Fortunately, some HCC-related lncRNAs are present in serum in a stable measurable form, and thus are easy to detect and analyze; hence they have a good potential to be used as novel biomarkers for HCC. However, the utility of most lnc-RNAs is still not fully investigated [17].
LncRNA urothelial carcinoma-associated 1 (UCA1), located on chromosome 19p13.12, was originally detected in bladder cancer, where its overexpression was associated with poor prognostic factors [15]. After-wards, it was found that it is upregulated in many other tumors such as stomach cancer, prostate cancer, breast cancer and HCC [18]. In the case of HCC, its expression was correlated with tumor size, stage, and vascular invasion [18-20].
Long non-coding RNAs wd repeat containing antisense to TP53 (WRAP 53) is an antisense transcript which controls the p53 tumor suppressor and was correlated with some head and neck tumors, breast cancer [21, 22] and HCC, where it was thought to have a role as an independent prognostic marker [19].
On the other hand, liver cirrhosis was reported to cause upregulation of some lncRNAs, and since in most cases HCC develops on top of liver cirrhosis, the reliability of lncRNAs in such a situation as a biomarker might be questionable [17]. Hereby, testing lncRNAs’ accuracy in differentiation between HCC and liver cirrhosis is crucial. This study was designed to investigate the potential role of UCA1 and WRAP53 in diagnosis of HCC as well as their ability in differentiation between liver cirrhosis and HCC.
Material and methods
Patients and study design
Sample size calculation was performed to detect an assumed area under the curve of 0.78 for using both markers to differentiate HCC from liver cirrhosis patients, with a significance level of 0.05 and 80% power; hence 90 subjects were recruited (30 patients diagnosed with HCC based on triphasic computed tomography (CT), 30 patients with liver cirrhosis, 30 healthy controls). They were recruited from the main University Hospital and Medical Research Institute Hospital, Alexandria University. Written informed consent was taken from all the patients and healthy controls. Patients with a history of active or cured other malignancies were excluded, as well as any patient with chronic inflammatory non-hepatic conditions. The study was conducted in accordance with the provisions of the World Medical Association Declaration of Helsinki. The study was reviewed and approved by Ethics Review Board of the Faculty of Medicine, Alexandria University (No. 0201516). The study has been registered in clinicaltrials.gov with clinical trial registration number NCT05088811.
History taking, clinical examination and ultrasound examination were done for all the subjects, and triphasic CT was performed in all patients with suspected/known HCC. Biochemical data including AFP, liver enzymes, serum albumin, bilirubin, and INR were collected.
Quantitative real-time PCR
For each subject, 10 ml of venous blood was obtained, followed by centrifugation for 10 min at 4°C. Cell-free serum was then stored at −80°C until RNA extraction.
Total RNA isolation from serum samples was performed using the miRNeasy Mini Kit (reference 217004, Qiagen, CA) The concentration and purity of RNA were measured using nanodrop then complementary deoxy ribonucleic acids (cDNA) were synthesized using the high-capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) according to the manufacturer’s recommendation. After that cDNA was stored at −20°C to be used in quantitative real-time polymerase chain reaction (RT-qPCR). RT-qPCR was performed on the Applied Biosystems Step-one Realtime PCR System using Thermo Scientific Maxima SYBR Green qPCR Master Mix (2X) (Thermo Scientific, USA) and specific primers for UCA1 and WRAP53.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The sequences of the PCR primers for UCA1, WRAP53 and GAPDH were as follows: UCA1, 5’-CTCTCCATTGGGTTCACCATTC-3’ (forward) and 5’-GCGGCAGGTCTTAAGAGATGAG-3’ (reverse). WRAP53, 5’-TG-GCACAAAGCTGGACAGT-3’ (forward) and 5’-GCTGGGTCCTGGTCTGAAG-3’ (reverse). GAPDH, 5’-GCACCGTCAAGGCTGAGAAC-3’ (forward) and 5’-TGGTGAAGACGCCAGTGGA-3’ (reverse).
The cycling conditions were 95°C for 10 minutes (initial cycle) followed by 40 cycles at 95°C for 15 seconds (denaturation) then annealing was done at 53°C for UCA1, 60°C for WRAP53 and 65°C for the GAPDH gene for 30 seconds then finally extension at 72°C for 30 seconds.
The fold change between a sample and a normal control for UCA1 and WRAP53 was calculated with the relative quantification method (RQ = 2–ΔΔCT).
Statistical analysis
Statistical analysis was done using IBM SPSS software package version 20.0 (Armonk, NY: IBM Corp.). Mann-Whitney and Kruskal-Wallis tests were used to compare quantitative variables. The serum levels of UCA1, WRAP53 and AFP were compared between the groups using the Kruskal-Wallis test, and pairwise comparison between each two groups was done using a post hoc test (Dunn’s test for multiple comparisons). Receiver operating characteristic (ROC) curves were used to evaluate the diagnostic value of serum UCA1, WRAP53 and AFP for HCC. A p-value of < 0.05 was considered statistically significant.
Results
Relative expression of serum UCA1 and WRAP53 in the three studied groups and their correlations with clinicopathological features of HCC
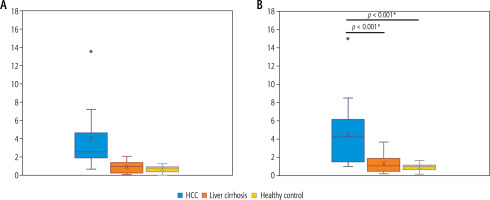
The median relative expression of serum UCA1 in patients with HCC was statistically significantly higher than that in patients with liver cirrhosis and healthy controls (2.62, 1.0 and 0.8 respectively) (p < 0.001). Serum WRAP53 relative expression was also higher in HCC patients than in patients with liver cirrhosis and healthy controls (4.25, 1.1 and 1.03 respectively) (p < 0.001). The difference in the relative expression of the two studied lncRNAs between patients with liver cirrhosis and healthy controls was not statistically significant (p > 0.05) (Fig. 1).
Fig. 1
Relative expression of serum UCA1 (A) and WRAP53 (B), both were significantly higher in patients with hepatocellular carcinoma (HCC) than in patients with liver cirrhosis and healthy controls
On correlation between serum UCA1 and different clinicopathological features of HCC, serum UCA1 overexpression was statistically significantly correlated with high serum AFP (defined as AFP more than 400 ng/ml) [23], presence of metastatic HCC, presence of lymph node metastases and the number of tumor foci being more than 3 (p < 0.05). On the other hand, high serum WRAP53 was significantly correlated with early stages of Barcelona classification of liver cancer (BCLC), defined as stages 0, A and B (p < 0.05). Serum WRAP53 was also statistically significantly higher in cases with no portal vein invasion and in cases where tumor size (defined as the size of the largest detected focal lesion) was less than or equal to 5 cm (p < 0.005) (Table 1).
Table 1
Correlation between serum UCA1, WRAP53 and clinicopathological features of hepatocellular carcinoma (HCC)
Receiver operating characteristic curve analysis for serum UCA1 and WRAP53 in HCC patients vs. patients with liver cirrhosis
ROC curve analysis was done to evaluate the ability of both markers to differentiate HCC from liver cirrhosis. For serum UCA1, area under the curve (AUC) was 0.9 (95% confidence interval [CI]: 0.82-0.98) and at the cutoff value of 2.1, the sensitivity and specificity were 73.3% and 100% respectively (p < 0.001). For serum WRAP53, AUC was 0.85 (95% CI: 0.76-0.94) and at the cutoff value of 2, sensitivity and specificity were 63.3% and 80% respectively (p < 0.001) (Fig. 2A).
Fig. 2
A) ROC curve for serum UCA1 and WRAP53 to differentiate between hepatocellular carcinoma (HCC) patients from patients with liver cirrhosis. B) ROC curve for combination of serum UCA1, WRAP53 and AFP to differentiate between HCC patients from patients with liver cirrhosis. C) ROC curve for serum UCA1 and WRAP53 to differentiate between HCC patients from patients with liver cirrhosis and healthy controls. D) ROC curve for combination of serum UCA1, WRAP53 and AFP to differentiate between HCC patients from patients with liver cirrhosis and healthy controls. E) ROC curve for serum UCA1 and WRAP53 to differentiate between HCC patients from healthy controls. F) ROC curve for combination of serum UCA1, WRAP53 and AFP to differentiate between HCC patients from healthy controls

ROC curve analysis was done to calculate the sensitivity and specificity on combining the two markers together or combining one of them with AFP or combination of all of them. The best results were achieved on combination of UCA1 with WRAP 53 and AFP (AUC = 0.97, 95% CI: 0.922-1.000, achieving sensitivity of 90% and specificity 100%) (p < 0.001) followed by adding of WRAP53 to AFP (AUC = 0.96, 95% CI: 0.915-1.000, achieving sensitivity of 86.7% and 93.3% specificity) (p < 0.001) (Fig. 2B).
Receiver operating characteristic curve analysis for serum UCA1 and WRAP53 in HCC patients vs. patients with liver cirrhosis and healthy controls
ROC curve analysis was also done to show the ability of the two markers to differentiate patients with HCC from the other two non-HCC groups (n = 60). For serum UCA1 area under the curve was 0.92 (95% CI: 0.86-0.98) and at a cutoff value of 1.5 it was found that it has sensitivity of 80% and specificity of 93.3% (p < 0.001). In the case of serum WRAP53, AUC was 0.9 (95% CI: 0.83-0.96) achieving sensitivity and specificity of 86.7% and 78.3% respectively, at a cutoff value of 1.3 (p < 0.001) (Fig. 2C).
On combination of the markers, the best results were also achieved on combination of serum UCA1 with WRAP53 and serum AFP (AUC = 0.982, 95% CI: 0.959-1.000, achieving sensitivity of 90% and specificity 100%) (p < 0.001), which was slightly better than adding serum WRAP53 to serum AFP (AUC = 0.979, 95% CI: 0.956-1.000, achieving sensitivity of 86.7% and 96.7% specificity) (p < 0.001) (Fig. 2D).
Receiver operating characteristic curve analysis for serum UCA1 and WRAP53 in HCC patients vs. healthy control
ROC curve analysis showed that AUC for serum UCA1 is 0.95 (95% CI: 0.89-1.00) and at a cutoff value of 1.28 it achieves sensitivity and specificity of 80% and 100% respectively. AUC for serum WRAP53 was almost like that of UCA1 (0.94, 95% CI: 0.80-1.00) with sensitivity of 86.7% and 93.3% sensitivity and specificity at cutoff value of 1.27 (p < 0.001) (Fig. 2E).
On combination of the two markers AUC is 0.99 achieving 90% sensitivity and 96.7% specificity, and on combining one or two markers with AFP, 100% sensitivity and specificity was achieved (p < 0.001) (Fig. 2F).
Receiver operating characteristic curve analysis for serum UCA1 and WRAP53 in HCC patients with negative AFP vs. patients with liver cirrhosis
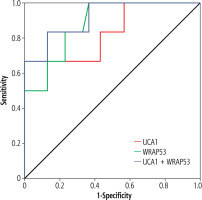
ROC curve analysis was performed for six cases in the HCC group who presented with negative AFP, defined as serum AFP < 20 ng/ml [24], and it was found that serum WRAP53 has a slightly better performance than serum UCA1, with AUC of 0.88 (95% CI: 0.73-0.96) and 0.83 (95% CI: 0.67-0.94) respectively. Serum WRAP53 at a cutoff value of 1.3 achieved 100% sensitivity, 63.3% specificity (p < 0.001) while serum UCA1 at a cutoff value of 2.1 achieved 66.7% sensitivity and 100% specificity (p < 0.01). Their combination has an AUC of 0.92 (95% CI: 0.79-1.00) achieving 83.3% sensitivity and 86.7% specificity (p < 0.001) (Fig. 3).
Discussion
Hepatocellular carcinoma usually develops on top of liver cirrhosis and usually is diagnosed in an advanced stage, which limits its management options; thus identifying a novel biomarker of high accuracy to detect early HCC is critically needed [8]. Alpha-fetoprotein is the commonly used biomarker for HCC; however, its performance as a surveillance tool is not satisfactory, mainly due to the high false positive rate [11-13].
LncRNAs are involved in the pathogenesis of various diseases, particularly malignant tumors, where they act as either oncogenes or tumor suppressor genes [15]. In addition, they have been identified as promising biomarkers for cancer detection due to their stability in body fluids and measurability [17, 18]. Several lncRNAs have been incriminated in oncogenesis of HCC and have been studied as potential diagnostic biomarkers for HCC. For instance, SCARNA10, HOTAIR, HULC and Linc00152 were found to be sig-nificantly higher in HCC and their combination with AFP improved their sensitivity and specificity [20, 25, 26].
In this study, we found that relative expression of serum UCA1 and WRAP53 was statistically significantly higher in HCC compared to patients with liver cirrhosis and healthy controls (p < 0.001); at the same time, UCA1 and WRAP53 expression did not have a statistically significant difference between patients with liver cirrhosis and healthy controls (p > 0.05). Kamel et al. [19] in their study that involved 82 HCC patients and 34 patients with chronic HCV infection and 44 healthy controls, found that both markers can discriminate between HCC and healthy controls but at significantly lower cutoff values than in our study (UCA1 of 1.04 vs. 1.28 in our study, WRAP53 of 0.61 vs. 1.27 in our study). However, their study did not directly test the ability of the two lncRNAs to discriminate between HCC patients and liver cirrhosis patients, which is from our point of view of critical importance since in most cases HCC develops on top of cirrhosis.
On the other hand, a single study performed to analyze the utility of eight different lncRNAs in HCC found that there was no significant difference of UCA1 level between HCC patients and healthy controls [20].
Using ROC curve analysis, the sensitivity and specificity of both markers were tested – alone and in combination with or without AFP – in various clinical scenarios; as in the case of differentiating HCC from liver cirrhosis, differentiating HCC from patients with liver cirrhosis and healthy controls and differentiating HCC from healthy controls. In the three scenarios, UCA1 alone performed slightly better in terms of sensitivity and specificity than WRAP53. On addition of AFP, the combination of the three markers has the best performance, achieving 100% specificity and 90-100% sensitivity, followed by the combination of WRAP53 and AFP achieving specificity ranging from 93% to 100% and sensitivity of 86% to 100% depending on the scenario of comparison.
This also moderately agrees with the results of Kamel et al. [19] in that the addition of both markers to AFP improved the overall sensitivity of HCC detection; however, their study found that this combination decreased the specificity to about 63% in contrast to our study that found that the combination of AFP with UCA1 and WRAP53 improved the specificity to about 97%. On the other hand, regarding UCA1 alone, our study strongly agrees with Zheng et al. [18] who found that sensitivity and specificity of UCA1 in differentiating HCC from liver cirrhosis was around 71% and 94% respectively with cutoff value of 1.99, which can be comparable to our findings of 73% sensitivity and 100% specificity at a cutoff value of 2.1. On comparing HCC to healthy controls, UCA1 achieved sensitivity of 73% and 99% specificity at a cutoff value of 1.85, which is comparable to our findings of sensitivity and specificity of 80% and 100% respectively at a cutoff value of 1.28.
On performing subgroup analysis for discriminating the HCC patients who were missed by AFP (< 20 ng/ml) from patients with liver cirrhosis, we found that WRAP53 has a slightly better performance than UCA1 with WRAP53 showing high sensitivity and moderate specificity and UCA1 showing the reverse with high specificity and moderate sensitivity and their combination achieving 83.3% sensitivity and 86.7% specificity. This agrees with Kamel et al. [19] who found that the addition of both markers to AFP in AFP negative patients increased the sensitivity of HCC detection significantly.
Generally, UCA1 seems to have the predilection to be correlated with poor prognosis, though the data on its correlation with different prognostic variables do not agree perfectly. Our study showed that UCA1 is correlated significantly with high AFP, presence of metastatic HCC, lymph node metastases and number of tumor foci, while there was no significant correlation between UCA1 and Child-Pugh class, portal vein invasion, BCLC stage or tumor size. A metanalysis that was performed to find the relation between UCA1 expression and lymph node metastasis with different types of tumors as HCC, colorectal cancer and other tumors revealed that a higher UCA1 expression predicted more lymph node metastasis and worse overall prognosis [27]. Another study showed that UCA1 is significantly correlated with advanced TNM stage and metastasis with no significant correlation between UCA1 tumor size and AFP [28].
Meanwhile, Zheng et al. [18] found that UCA1 expression was significantly correlated with tumor size, vascular invasion and TNM stage, while at the same time there was no statistically significant correlation with high AFP (defined as AFP ≥ 400 ng/ml) or number of foci or presence of metastases. The correlation between UCA1 and tumor size and vascular invasion was also observed in a previous study performed on tissue UCA1 [29]. Kamel et al. [19] found a significant association of UCA1 and Child-Pugh score and noted that its expression was higher in those cases with large tumor size but did not reach statistical significance.
On the other hand, WRAP53, in our study, was found to be statistically significantly higher in the case of early stages of HCC (defined as BCLC 0, A or B), smaller size of focal lesion (< 5 cm) as well as the absence of portal vein invasion, which suggests that WRAP53 might have the utility to detect early stages of HCC. However, this partially disagrees with the findings of Kamel et al. [19] who found a significant association between WRAP53 and advanced Child-Pugh score.
The oncogenic mechanism of most lncRNAs is not yet fully elucidated. UCA1 may have a role in inhibiting some tumor suppressors such as miR-216b [28], miR-203 [29], miR-184, miR-204-5p and miR-182 [18]. WRAP53 has a role as an antisense transcript of the P53 gene and as a subunit of telomerase. It was found that knockdown of the WRAP53 protein induces apoptosis, especially tumor cells as compared with normal human cells, indicating that cancer cells depend on WRAP53 expression for their survival [21]. However, the specific role of WRAP53 in HCC development is not yet studied.
Conclusions
Serum UCA1 and WRAP53 are overexpressed in HCC and could be used alone, or in combination or with AFP, as diagnostic non-invasive biomarkers for HCC with accepted sensitivity and specificity. Interestingly, WRAP53 expression seems to have the potential to detect early BCLC stages of HCC, while UCA1 expression is correlated with advanced stages and poor prognostic signs such as metastasis and vascular invasion. Both markers might have the potential to be used to detect AFP negative HCC. However, this study is limited by the small sample size. Further multicentric studies are needed to confirm these findings and to clarify their role in tumorigenesis of HCC.