Introduction
Cutaneous squamous cell carcinoma (SCC), widely known as epidermoid carcinoma, is the second most common form of skin malignancy, representing around 20% of all skin cancers [1]. In the United States, cutaneous SCC is the main cause of death due to non-melanoma skin cancer each year [2]. The incidence of cutaneous SCC mainly develops from changes in the epidermal homeostasis resulting in uncontrolled proliferation of epidermal cells [3]. In the current clinical setting, surgical resection remains the primary treatment for in situ cutaneous SCC. However, it is not suitable for some patients due to its invasiveness. For instance, tumours at critical sites such as the eyelids require non-surgical management for tissue preservation [4]. Moreover, metastases responsible for cancer advancement are associated with poor outcome in metastatic SCCs [5]. Given the high frequency of metastatic SCCs on a global scale and poor prognosis for advanced stages, scientists are inventing better techniques in search of non-invasive approaches to cure cutaneous SCC more effectively.
It is well documented that the secretion of matrix metalloproteinase (MMP) proteases is crucial in tumor angiogenesis, invasion, and metastasis [6]. Invasion of neoplastic cells into adjacent stromal regions is assisted by MMPs through the degradation of neighbouring extracellular matrix. The role of MMPs in skin cancer is well-established. Among others, matrix metallopro-teinase 9 (MMP9 ) is notably responsible for tumour vascularisation [5, 7]. Matrix metallopro-teinase 9 is a type IV collagenase, which plays a major role in the metastatic spread of tumours by digesting the basement membrane type IV collagen. This potent proteolytic enzyme is key target of therapeutics focused on preventing vascular invasion and cancer metastasis. Previously, researchers reported the use of various methods to inhibit the activities of MMP9 in the effort to suppress its influence on malignant development and invasion [8–12]. Small interfering RNA (siRNA) was used to knockdown tissue transglutaminase resulting in downregulated cell secretion of MMP9 [11]. It was noted that invasion and attachment of cutaneous SCC cell line A431 were inhibited. Treated with bioactive compound rhodomyrtone naturally synthesised from Rhodomyrtus tomentosa leaves, cutaneous SCC cells showed lowered protein expression of MMP9, and consequently suppressed metastatic activities [12]. Although these studies suggested novel anti-metastasis therapies, the suppression of MMP9 was not direct and immediate. Targeted inhibition of MMP9 at the genetic level should be initiated to treat skin cancer more effectively.
This study aimed to determine whether the MMP9 gene is a suitable gene target for anti-cancer therapy for cutaneous SCC. As such, the present study was designed to evaluate the effect of inhibition of MMP9 gene on cell viability and metastatic capability of cutaneous SCC cell line through clustered, regularly interspaced short palindromic repeat (CRISPR) transfection of guide RNA (gRNA) targeting the MMP9 gene.
Material and methods
Cell culture
The human cutaneous SCC cell line A431 was obtained from the UPM-MAKNA Cancer Research Laboratory, Institute of Bioscience, Universiti Putra Malaysia. The cells in cryovial were revived and cultured in a 75 cm2 culture flask with 10 mL of Dulbecco’s Modified Eagle Medium (DMEM, Gibco, USA) supplemented with 10% Foetal Bovine Serum (FBS) (Gibco, USA) and 1% penicillin/streptomycin (Gibco, USA). The flask of cells was incubated in a humidified incubator supplied with 5% carbon dioxide at 37°C. The cells were expanded by performing subculture whenever the cells achieve 80% confluency. Trypsinization was performed by incubating the cells with 2.5 g/l trypsin- ethylenediaminetetraacetic acid (Nacalai Tesque Inc, Kyoto, Japan) for 10 min, after which complete culture medium was added and centrifuged at 180 × g for 5 min. Fresh medium was added to the resulting pellet and the cells were distributed to new sterile culture flasks.
Transfection of guide RNAs into A431 cell line
The design of guide RNAs
With the use of CRISPR design tool by Integrated DNA Technologies (IDT, Singapore), 2 gRNAs were designed and synthesised for the target gene MMP9. The sequences of the gRNAs are listed in Table 1.
Transfection of guide RNA into A431 cell line
The inhibition of MMP9 gene was carried out by transfecting the gRNAs and Cas9 protein (IDT, Singapore) into A431 cells. Ribonucleoprotein (RNP) complex was formed by mixing equimolar (ratio 1 : 1) of a gRNA and Cas9 protein. Briefly, in each well, 16.5 µl of 1 µm gRNA and 16.5 µl of 1 µm Cas9 protein were added to 17 µl of Opti-MEM medium (Gibco, USA). The mixture was then resuspended and incubated at room temperature for 10 min. After that, 1.2 µl of jet CRISPR (Polyplus-transfection, France) was added to the mixture, resuspended, and incubated at room temperature for 15 min. The transfection solution was prepared in a reservoir before being distributed into each well. Then, 1 × 105 A431 cells in 500 µl medium was seeded to each well. The plate was then incubated at 37°C for 48 hours.
Determination of transfection efficiency
Following 48 hours of transfection, the expression level of the MMP9 gene in A431 cells was assessed by polymerase chain reaction (PCR). The total RNA was isolated using RNeasy Mini Kit (Qiagen, USA) and converted to cDNA with SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific, USA). Polymerase chain reaction was carried out using GoTaq Green Master Mix (Promega, USA) with primers: (forward primer: 5-TTCCAAACCTTTGAGGGCGA-3 and reverse primer: 5-CAAAGGCGTCGTCAATCACC-3) [13]. Quantification of PCR product concentration and purity were measured using NanoDrop UV Spectrometer (PCR max Lambda, USA). The polymerase chain reaction products were then separated in 1.8% agarose gel with EtB “Out” Nucleic Acid Staining Solution (Yeastern Biotech, Taiwan) at 90 V for 45 min. Gels were photographed using the G: box gel documentation system (Syngene, UK). The intensity of the bands specific to MMP9 gene were normalised with that of housekeeping gene β-actin, and then compared between groups, using ImageJ software (Maryland, USA) to determine the transfection efficiency.
Cell viability assay
Acridine orange/propidium iodide staining
To visualise the cell viability of A431 cells after 48 hours of CRISPR transfection, the cells were trypsinised and centrifuged to obtain cell pellets. For each sample, 10 µl of cell pellet was added with 10 µl of staining solution, which consists of equal volumes of acridine orange (AO) (10 µg/ml) and propidium iodide (PI) (10 µg/ml). The stained cell solution was dropped onto a clean glass slide and covered with a coverslip before being viewed under a fluorescence microscope within 30 min.
MTT assay
The 3-[4, 5-methylthiazol-2-yl]-2, 5-diphenyl-tetrazolium bromide assay (MTT, Ruibio, Zhejiang, China) was used to quantitate A431 cell viability after the inhibition of MMP9 gene. After cell incubation in CRISPR transfection reagents for 48 hours, the reagents were discarded and 40 µl of MTT reagent added into each well following by incubation at 37°C for 4 hours. Then, the reagent was discarded and 100 µl of dimethyl sulfoxide was added into each well and incubated at 37°C for 10 min. The absorbance reading of the samples was quantified using a micro-plate reader (Bioscience, USA) at the wavelength of 570 nm. The reading at reference wavelength of 650 nm was used for normalisation.
Cell migration assay
The scratch assay was conducted to study the metastatic capacity of cells after CRISPR transfection. In the middle of A431 cell monolayer, a scratch wound was created using a sterile 200 µl micropipette tip. After rinsing gently with 1× phosphate buffer saline (PBS, Gibco, USA), transfection reagents were added to the cells. The wounded cell monolayer was observed at 0–48 hours using an inverted phase- contrast microscope at 100× total magnification. The wound width was measured by ImageJ software (Maryland, USA). The rate of cell migration was calculated by the formula:
RM = (Wi – Wf)/t
RM – rate of cell migration (µm/h), Wi – initial wound width (µm), Wf – final wound width (µm), t – duration of migration (h)
Quantitative polymerase chain reaction
The expression levels of cancer-promoting genes were assessed following 48 hours of transfection. The total RNA was isolated using RNeasy Mini Kit (Qiagen, USA) and converted to cDNA with the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific, USA). Thunderbird SYBR quantitative polymerase chain reaction (qPCR) Mix (Toyobo, Japan) was used to perform qPCR in a Light Cycler 480 real-time PCR System (Roche Molecular Systems, California, USA). The mRNA expression of cancer-promoting genes were evaluated by their primers: TGF-β (forward primer: 5-CTGTCCAACATGATCGTGCG-3 and reverse primer: 5-GACACAGAGATCCGCAGTCC-3), FGF (forward primer: 5-TCCACCTATAATTGGTCAAAGTGGT-3 and reverse primer: 5-CATCAGTTACCAGCTCCCCC-3), PI3K (forward primer: 5-GGTGAAGCTCGTGTGTGGA-3 and reverse primer: 5-GAAGACAGGGCTCCACTTCC-3), VEGF-A (forward primer: 5-GGGAAAGGGGCAAAAACGAA-3 and reverse primer: 5-AGGCTCCAGGGCATTAGACA-3), vimentin (forward primer: 5-GTACGACTATGGCGCTCTGG-3 and reverse primer: 5-TGATGGCACTCTGGAGCTTG-3). The threshold cycle values were normalised with housekeeping gene β-actin (forward primer: 5-GACAGGATGCAGAAGGAGATTACTG-3 and reverse primer: 5-CTCAGGAGGAGCAATGATCTTGAT-3), and the relative fold change in gene expression was calculated by 2-ΔΔCt method.
Statistical analysis
The experiment results were expressed as mean values ± standard error of the mean (S.E.M). Prism version 8.0 statistical software (GraphPad, Software Incorporated San Diego, CA) was used to perform one-way analysis of variance (ANOVA) and post hoc Tukey’s multiple comparison test. Results were considered as statistically significant when p-values were below 0.05.
Results
Reduced mRNA expression of MMP9 gene after clustered regularly interspaced short palindromic repeats transfection
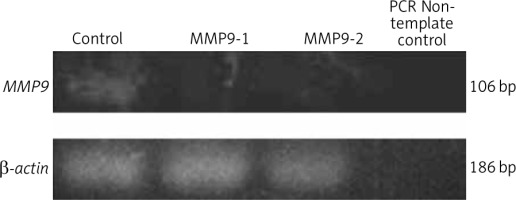
Polymerase chain reaction was conducted to amplify the MMP9 gene in A431 cells following CRISPR transfection. The gel electrophoresis results of the PCR products showed less intense bands for CRISPR transfected samples, i.e. MMP9-1- and MMP9-2-transfected A431 cells (Fig. 1). Analysis of band intensity using ImageJ software revealed that the expression of MMP9 gene in MMP9-1- and MMP9-2-transfected cells was downregulated by 95.3% and 95.7%, respectively, as compared to non-transfected A431 cells.
Reduced viability of cutaneous squamous cell carcinoma cells after treatment
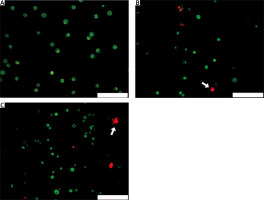
Acridine orange/propidium iodide double staining of A431 cells was performed to visualise the effect of CRISPR transfection of gRNA targeting the MMP9 gene on cell viability. After 48 hours of CRISPR transfection, the viable cells were stained green by AO dye, while apoptotic cells were stained red by PI dye. Under fluorescence microscope, non-transfected A431 cells were all stained green (Fig. 2 A), indicating that they were all live cells, whereas CRISPR transfected A431 cells showed green and red cells (Fig. 2 B, C); this indicated that after 48 hours of CRISPR transfection, some cells experienced apoptosis and had reduced cell viability. The morphological characteristics of apoptotic cells were also observed, including membrane blebbing and appearance of apoptotic bodies.
Fig. 2
Acridine orange/propidium iodide double staining of A431 cells after 48 hours of clustered, regularly interspaced, short palindromic repeat transfection. Only green cells were observed in nontransfected cells (A), MMP9-1- (B) and MMP9-2-transfected cells (C) showed morphological feature of apoptosis, including membrane blebbing and collapse of cells into apoptotic bodies (white arrows)
The scale bars denote 200 µm
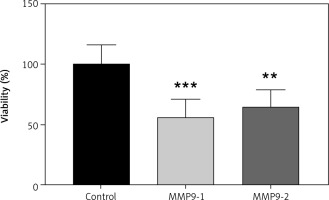
MTT was used to quantify A431 cell viability after transfection of gRNA targeting on the MMP9 gene. After 48 hours of CRISPR transfection, the cell viability of A431 cells was calculated and normalised with non-transfected cells. The data are presented as a bar graph in Figure 3. The cell viability of A431 cells have reduced to 56.27% (p < 0.001) and 65.29% (p < 0.01) following 48 hours of CRISPR transfection of gRNAs MMP9-1 and MMP9-2, respectively.
Fig. 3
Quantitative assessment of A431 cell viability by MTT assay after 48 hours of clustered, regularly interspaced, short palindromic repeat transfection
The inhibition MMP9 genes reduced cell viability A431 cells. The data were expressed as mean values ± S.E.M (n = 9). Statistical analysis using one-way analysis of variance and post hoc Tukey’s multiple comparison test was performed. Non-transfected cells were used as a control group.
** p < 0.01
***p < 0.001
Reduced migratory activity of cutaneous squamous cell carcinoma cells after treatment
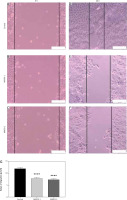
A scratch assay was carried out to evaluate the A431 cell capacity of cellular migration following CRISPR treatment. Non-transfected A431 cells displayed excellent migratory activity and closed the wound gap faster than all the transfected cells (Fig. 4 D). Meanwhile, other transfected A431 cells showed lower potential to migrate and heal the wound after 48 hours of CRISPR transfection (Fig. 4 E, F). The wound width was measured and calculated by software ImageJ. The rates of cell migration were reduced significantly in A431 cells after CRISPR transfection of gRNA targeting the MMP9 gene, relative to that of non- transfected cells (Fig. 4 G).
Measurement of width of scratch gap was performed using ImageJ software. The rates of cell migration for non-transfected cells were 11.62 µm/h, while the rates for MMP9-1- and MMP9-2-transfected cells were 7.73 µm/h and 7.25 µm/h, respectively. The results were significant (p < 0.0001) in both experimental groups compared with the control group (Fig. 4 G).
Fig. 4
Scratch assay of A431 cells after clustered, regularly interspaced, short palindromic repeat (CRISPR) transfection for 48 hours. A scratch wound was created in the middle of the cell monolayer at the time of CRISPR transfection (A–C), non-transfected cells showed smallest wound gap after 48 hours (D), MMP9-1- and MMP9-2- transfected cells showed less migratory activity after 48 hours (E, F), rate of cell migration after 48 hours of CRISPR transfection (G)
The scale bars denote 200 µm. The rates of cell migration were reduced in both transfected groups compared to non-transfected cells. The data are expressed as mean values ± S.E.M (n = 9). Statistical analysis using one-way analysis of variance and post hoc Tukey’s multiple comparison test was performed.
**** p < 0.0001
Changes of mRNA expression of cancer-promoting genes after treatment
Quantitative PCR analysis was performed to find out the changes in cancer-promoting gene expression in A431 cells after CRISPR transfection of gRNA targeting the MMP9 gene. All the data were normalised to housekeeping gene β-actin, and the results were calculated using comparative CT method (2-ΔΔCt).
TGF-β, FGF, and PI3K are involved in cancer cell proliferation. Following CRISPR transfection of MMP9-1 and MMP9-2 into A431 cells, the mRNA expression levels of TGF-β were downregulated 1.23- and 0.25-fold, respectively (Fig. 5 A), whereas the mRNA expression levels of FGF were downregulated 2.02-fold and 1.88-fold, respectively (Fig. 5 B), MMP9-1-transfected cells were downregulated PI3K 0.89-fold, and the expression was upregulated 0.05-fold in MMP9-2-transfected cells (Fig. 5 C).
Fig. 5
Differential expression of genes involving in cancer cell proliferation and metastasis in A431 cells after 48 hours of clustered, regularly interspaced, short palindromic repeat transfection. Relative fold changes of gene expression of TGF-β (A), FGF (B), PI3K (C), VEGF-A (D) and vimentin (E). Results were calculated using comparative CT method (2–ΔΔCt) where β-actin was used to normalise the threshold cycle values obtained from qPCR. Data are displayed as mean log2 fold change ± S.E.M (n = 3). Statistical analysis was performed using Mann-Whitney U test between the control group (non-transfected cells) and each of the experimental groups (transfected cells) individually
* p < 0.05
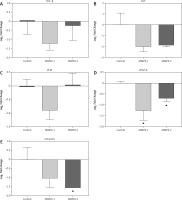
VEGF-A and vimentin are epithelial to mesenchymal transition markers responsible for cancer cell invasion and metastasis. Significantly downregulated expressions of VEGF-A gene expressions were decreased 1.27- and 0.7-fold in MMP9-1 and MMP9-2 transfected cells, respectively (p < 0.05) (Fig. 5 D).
Downregulated expressions of vimentin was observed in both MMP9-1 and MMP9-2 transfected groups, with 1.02- and 1.54-fold reduction, respectively. The fold change in vimentin gene expression was found to be significant in MMP9-2 but not MMP9-1 transfected cells (Fig. 5 E).
Discussion
The tumoural expression of protease MMP9 was remarkable during various types of cancer advancement to an aggressive phenotype [14]. During carcinogenesis of cutaneous SCC, tissue remodeling and inflammation were also prominent with elevated expression of MMP9 [15]. Upregulated MMP9 expression in keratinocytes in advanced carcinomas could induce more rapid degradation of the extracellular membrane, thus facilitating tumour invasion to neighbouring tissues [14]. The enhanced expression level of MMP9 has also been associated with poor prognosis in cancer patients.
We hypothesised that direct inhibition of the MMP9 gene would suppress cancer activity of cutaneous SCC cells. In this study, after 48 hours of CRISPR transfection of gRNA targeting the MMP9 gene into cutaneous SCC cell line, both the viability and migratory activities of treated cells were reduced significantly in comparison to the non-transfected cells (Fig. 3, 4). Apoptosis was also observed after CRISPR transfection of gRNA targeting the MMP9 gene (Fig. 2), which could be attributed to the consequences of the transfection protocol [16].
Analysis of mRNA levels of important cancer-promoting genes revealed relatively low expression in CRISPR transfected cells as compared to non-transfected cells. Tumour cells metastasise through the formation of new vasculatures via a process known as angiogenesis. A major marker of angiogenesis, VEGF-A, was markedly downregulated after CRISPR transfection (Fig. 5), indicating inhibited metastasis. A previous study showed that targeting MMP9 could reduce the density of microvessel and inhibit angiogenesis in tumours [17]. Vimentin is also responsible for migratory activities in cancer cells through the formation of lamellipodia and invadopodia during tumour invasion. Previous reports have shown defective migration in tumour cells with loss of vimentin [18, 19]. In oral SCC cells, lower levels of vimentin were found in non-metastatic cells when compared to the metastatic phenotype [20].
While significant downregulation of VEGF-A and vimentin noted in the transfected cells in this study indicated that the progression of cutaneous SCC cells to the metastatic phenotype was successful inhibited, the expressions of oncomarkers TGF-β, FGF, and PI3K were not downregulated significantly in this study (Fig. 5). This indicated that MMP9 plays a more important role in cancer cell metastasis compared to cancer cell viability and proliferation. Similarly, results from scratch assay (Fig. 3 G) were also more significant (p < 0.0001) than the results from MTT assay (Fig. 2). In future studies, gene expression analysis of other MMP9-associated genes can be included to assess the impact of MMP9 gene knockdown on their expression.
CRISPR-Cas9 gene editing technique is sequence-specific because it targets on the DNA level [21, 22]. It supports direct cleavage of complementary DNA sequences and produces double-stranded breaks (DSB). The created DSB will be inherently corrected by random nucleotide insertion and deletion and ultimately result in stop codon [23]. Consequently, transcription of the target gene is inhibited. In this study, we harnessed this prokaryotic immunity system to target MMP9 genes in cutaneous SCC, by transfecting MMP9-specific gRNAs into A431 cell line. After CRIPSR transfection, the mRNA expression level of the MMP9 gene was downregulated (Fig. 1).
Recently, CRISPR-Cas9 genome editing was successfully used for the first time to treat a patient with Leber congenital amaurosis type 2, a type of inherited eye disorder causing vision loss at birth, by direct intravenous administration [24]. Besides being less invasive, specific CRISPR gRNA is simple to design. Furthermore, significant results were obtained in only 48 hours in the present study. In vivo and clinical studies should be initiated for CRISPR-based anti-cancer therapy in the future.
In addition, the design of the experiment could be improved to target MMP9 more effectively. Co-transfection of 2 or more gRNAs targeting MMP9 should be performed in future studies to improve transfection efficiency in order to achieve better anti-cancer results. Recently, researchers have proven that concurrent transfection of five gRNAs in vitro is possible with CRISPR-Cas9 transfection, to achieve high gene editing efficiency [26]. Previously, we have reported reduced migratory activities in A431 cells following CRISPR transfection of gRNA targeting on MMP2 [27]. It has also been demonstrated that multiple genes could be targeted using a single CRISPR system in a simultaneous transfection event [22]. Simultaneous targeting of both MMP2 and MMP9 could deliver more favourable outcomes in inhibiting cutaneous SCC.
Conclusions
In conclusion, the current study demonstrated that MMP9 is involved in cutaneous SCC cell survival and metastasis, with a more important role in metastasis. Clustered, regularly interspaced short palindromic repeats mediated inhibition of MMP9 gene suppressed the survivability and migratory capacity of cutaneous SCC cells. Clustered regularly interspaced short palindromic repeat treatment serves as a potential non-invasive approach directly targeting on the DNA level of oncogenes, for the management of skin cancer and other types of metastatic malignancies.