Introduction
The tumor microenvironment and the population of tumor cells undergo rapid changes. Genetically distinct mechanisms of resistance towards the epidermal growth factor receptor (EGFR) blockade may arise in different metastases, constituting another challenge for the efficacy of targeted therapy [1]. Examination of circulating tumor DNA (ctDNA) allows tracking of mutation status in real time and reflects all cancer cell populations. On the other hand, low tumor DNA content in plasma may cause failure of liquid biopsy to detect any mutation in crucial genes [1]. Acquired EGFR c.2369C>T p.Thr790Met (T790M) mutation is detected in up to 69% [2] of cancers resistant to the first-generation tyrosine kinase inhibitors [3]. However, other potential resistance mechanisms, such as the mutation EGFR p.Cys797Ser [4], mutations in EGFR in codon 792 [5] or the recently reported EGFR p.Leu718Val (conferring resistance to osimertinib, but with retained susceptibility to afatinib) [6], indicate the importance of genomic diagnostics and the need for screening of a broad spectrum of mutations to better discover mechanisms of resistance.
ctDNAs present in blood are fragmented, and therefore their stability is limited. According to studies that compare the utility of different blood collection tubes (BCT) – tubes with K2EDTA or K3EDTA anticoagulants, tubes with stabilizers for white blood cells and cell free DNA (cfDNA) (Streck, CellSave) – those with K2EDTA or K3EDTA can be used for ctDNA analysis when the period between blood collection and plasma separation is sufficiently short [7–9]. Monitoring of the ctDNA mutation status in blood can help with detection of residual disease, recurrence, relapse and resistance [10].
Material and methods
The enrolled patients (1 male and 6 females) were recruited at the Franciszek Łukaszczyk Oncology Centre and granted their informed consent for mutation testing. All patients were Caucasian and were diagnosed with metastatic adenocarcinoma of the lung. The patients’ median age at the start of gefitinib, erlotinib or afatinib therapy was 69 years. Assessment of the EGFR status was performed in the period 2012–2015 based on DNA isolated from adenocarcinoma cells derived from formalin-fixed paraffin-embedded (FFPE) tissue samples (3 patients) or cytology samples (4 patients). Representative biopsy or FFPE samples with a range of 10–80% of tumor nuclei were selected by pathomorphologists (Table 1). Samples identified with a small percentage of tumor cells had to meet previously described criteria [11]. DNA isolated from FFPE samples was extracted using the FFPE QIAamp DNA Tissue Kit (QIAGEN). EGFR mutation detection was performed using the CE-IVD EGFR Mutation Analysis Kit (Entrogen). ctEGFR analysis was conducted for 20 plasma samples derived from 7 enrolled patients. For ctDNA detection, 4 ml blood samples were collected into regular blood collection tubes at 6-month intervals and immediately centrifuged for plasma collection. Time from the moment of blood collection to the start of centrifugation to separate plasma was 2–12 minutes for all 20 samples. The amount of plasma from which ctDNA was isolated ranged from 1,050 to 2,400 μl. Plasma samples were stored at –86°C until used. ctDNA was isolated using the QIAamp Circulating Nucleic Acid Kit (QIAGEN). All 20 DNA samples derived from liquid biopsies passed the internal quality control for circulating tumor EGFR mutation analysis. Simultaneous detection of the most common mutations in exons 19, 20 and 21, including the inhibiting p.Thr790Met mutation in EGFR, in the ctDNA extracted from the patients’ plasma was performed using the ctEGFR Mutation Detection CE-IVD kit (Entrogen).
Table 1
Characterization of the group of patients qualified for TKI therapy
| No. | Gender | Age* | Content of tumor nuclei (%) | Mutation detected | TKI | ctEGFR | Progression/ Stabilization | |
|---|---|---|---|---|---|---|---|---|
| FFPE | Cytology | |||||||
| 1 | F | 77 | 25 | N/A | Ex19del | Erlotinib | No mutation No mutation No mutation | Stabilization Stabilization Stabilization |
| 2 | F | 83 | 70 | N/A | Ex19del | Gefitinib ** | No mutation No mutation T790M | Stabilization Stabilization Progression |
| 3 | F | 82 | 80 | N/A | L858R | Gefitinib ** | L858R T790M L858R T790M L858R T790M | Stabilization Progression Progression |
| 4 | M | 55 | 10 | N/A | Ex19del | Afatinib ** | Ex19del Ex19del | Stabilization Progression |
| 5 | F | 68 | 20 | Ex19del | N/A | Afatinib | No mutation No mutation No mutation | Stabilization Stabilization Stabilization |
| 6 | F | 53 | 10 | Ex19del | N/A | Erlotinib ** | No mutation No mutation No mutation | Stabilization Stabilization Progression |
| 7 | F | 68 | 80 | Ex19del | N/A | Afatinib | wEx19del T790M No mutation | *** Stabilization Stabilization; 1 year later – brain metastasis |
Results
The average time from blood collection to plasma processing at the Department of Molecular Oncology and Genetics of the Oncology Centre in Bydgoszcz was approximately 6 minutes and 30 seconds. Instead of blood storage at 4°C, we immediately prepared plasma for molecular analysis and were able to detect ctEGFR mutation in the sample with the lowest plasma collection volume (1050 μl).
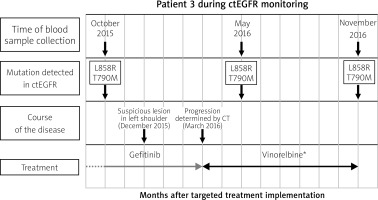
The most common somatic deletions in exon 19 and one p.Leu858Arg mutation in exon 21 of the EGFR gene were detected (Table 1). In 6 out of 7 cases, the first assessment of the ctEGFR status in plasma was performed in patients who had already started therapy with TK inhibitors, and patient 7 had an exon 19 deletion detected in ctDNA before targeted treatment implementation. During the 15-month plasma collection and ctEGFR mutation monitoring period, stabilization of the condition of two patients, no. 1 and 5, during TKI treatment coincided with a lack of p.Thr790Met mutation detection in ctEGFR (Fig. 1). Repeated detection of the p.Thr790Met mutation together with the L858R mutation was found in patient 3 (Fig. 2). At the time of the first p.Thr790Met detection in ctDNA, the patient was described as clinically stabilized. Two months later, a suspected lesion in the left shoulder was detected. Clinical progression was confirmed 4 months after the first p.Thr790Met detection in plasma (Fig. 2). Other single detections of the p.Thr790Met mutation were found in plasma derived from patients 2 and 7 followed by progression confirmed by X-ray (patient 2) and CT (patients 2 and 7).
Fig. 1
Example of qPCR mutation analysis in the EGFR gene in DNA isolated from an FFPE tissue sample (I) and ctDNA in patient 1 (II.A, II.B, II.C). Deletion in EGFR exon 19 (I; July 2015) detected in FFPE cancer tissue. No EGFR mutation detected in plasma ctDNA in November 2015 (dotted arrow) – II.A, May 2016 – II.B, December 2016 – II.C. Stabilization of disease progression. Lack of detection of exon 19 deletion in ctEGFR from November 2015 to December 2016 correlates with stabilization of disease confirmed clinically using a broad range of medical imaging techniques
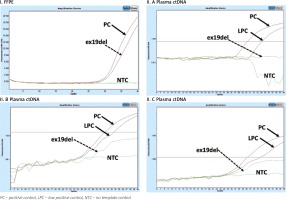
Fig. 2
Example of monitoring ctEGFR mutation in patient during TKI treatment. The X-axis shows the time from the beginning of targeted therapy, the Y-axis refers to the detection of mutation. Blood samples were collected 3 times: October 2015, May 2016 and November 2016. Despite clinical stabilization, p.Thr790Met mutation was detected in October 2015 (2 months before appearance of a suspicious lesion in the left shoulder and 3 months before clinical confirmation of metastasis in scapula determined by CT). * In accordance with the fact that osimertinib TKI was introduced to Polish healthcare from November 1, 2017, the patient monitored in 2015–2016 received the next treatment line consistent with the guidelines of the Polish Health Ministry at that time
Discussion
Over the years, the analysis of somatic mutations has become a standard procedure performed using postoperative material or biopsy. Currently, new sources of tumor DNA, such as blood [12], saliva or cerebrospinal fluid, have become available (reviewed in [13]). This noninvasive approach allows identification of ctDNA and monitoring of tumor dynamics and genetic alterations at advanced stages of the disease. The half-life of tumor DNA is estimated at 16 min, based on the data obtained from the clearance of fetal DNA in maternal plasma [13], which translates into approximately 6 × 108 mutant fragments released from the tumor every day. For patients with a tumor load of 100 g in size (approximately 3 × 1010 neoplastic cells), Diehl et al. estimated that 3.3% of the tumor DNA is released into the circulation on a daily basis. It is known that blood tubes, variations in storage conditions, different transportation methods and processing into plasma [13, 14] influence sample degradation and often have a negative impact on the ctDNA test performance and accessibility. In an interesting study, Parpart-Li [15] employed new generation sequencing (HiSeq, Illumina) to detect and compare mutations in plasma isolated from K2EDTA and BCT tubes (Streck). They used a targeted panel to sequence 58 genes which covered over 80 thousand nucleotides. The obtained base error rate (calculated as the number of mutated bases divided by the number of targeted bases) was similar and did not show any statistically significant difference (p = 0.28) between plasma isolated from K2EDTA and BCT tubes. On the other hand, simple steps, such as immediate plasma preparation or K2EDTA storage at 4°C, minimize the release of cfDNA by cells undergoing lysis or necrosis [15].
As preanalytical variables for ctDNA including specimen collection and handling are well understood [13, 14, 16] we did not perform direct comparison of ctDNA mutations detection isolated from K2EDTA and BCT tubes but put attention to immediate plasma separation (average time 6 min 30 s).
The p.Thr790Met mutation can also be assessed at academic research centers and commercial laboratories in Poland using standard collection of peripheral blood into K2EDTA tubes and isolation of plasma by centrifugation within 3 hours [17]. Reference oncology centers, such as the Franciszek Lukaszczyk Oncology Centre in Bydgoszcz, in which procedures such as transportation, storage and immediate processing of biological material are standardized, follow restrictive rules and take from 2 to 12 minutes. Our findings demonstrate the usefulness of the more cost-effective method employing the cheaper K2EDTA-coated tubes instead of the BCT Streck tubes. However, looking at this preanalytical process from a different perspective, local and regional laboratories which send blood to be processed at reference oncology centers should be obligated to use cell-free DNA BCT Streck tubes in order to decrease the influences on that process.
Another question assessed in this study was whether liquid biopsy could be a good molecular monitoring tool. As a biomarker, we chose circulating tumor DNA derived from adenocarcinoma patients treated with TK inhibitors. The first important element of the monitoring of the TK inhibitor efficacy using genetic methods is cooperation between clinicians and geneticists. This is due to the preclinical process described above and the need for a well-planned material collection, processing and testing procedure. Our experiment showed that not all patients treated with TKIs agreed to be enrolled in the study (data not shown), but the majority (6 out of 7) of the enrolled ones donated blood three times at 6-month intervals. We did not detect a ctEGFR mutation in 2 out of 7 patients who were under erlotinib (patient 1) or afatinib (patient 5) treatment. However, patients were clinically stabilized at the time of the study. In the other two patients who demonstrated progression during the evaluation period, EGFR ex19 deletion was only detected in plasma ctDNA derived from patient 4, while none of the tested mutations was detected in plasma ctDNA derived from patient 6. Failure of the ctEGFR p.Thr790Met detection could be due to three reasons: isolation of ctDNA from an insufficient amount of plasma (blood collected into 4 ml K2EDTA tubes) in comparison with regular blood collection for liquid biopsy (10 to 20 ml), use of qRT-PCR instead of ddPCR or BEAMing methodology, or development of resistance mechanisms to afatinib other than p.Thr790Met by patient 4. Finally, p.Thr790Met was detected in ctDNA in 3 out of 7 patients. All of them demonstrated progression of the disease: patients 2 and 3 – progression of lung lesions determined by CT in less than 4 months (Fig. 2); and patient 7 – brain metastases found 2 years after the p.Thr790Met detection. Unfortunately, we were not able to compare the presence of p.Thr790Met in ctDNA and in metastatic lesions. It needs to be mentioned that the intermediate sensitivity of plasma genotyping for the detection of p.Thr790Met has been reported at 70%, and the plasma p.Thr790Met-negative population is a mixture of true and false negatives; therefore biopsy is needed to further investigate the presence of p.Thr790Met-positive tumor tissue [18, 19]. Updated in 2018, molecular testing guidelines for the selection of lung cancer patients for TKI therapy recommend firstly a tissue sample for analysis, but if tissue biopsy material is unavailable or insufficient and tissue rebiopsy is not feasible, then a ctDNA assay for an EGFR mutation may be performed [19]. Finally experts had a consensus opinion that clinicians may use plasma ctDNA methods to evaluate EGFR p.Thr790Met mutation in lung adenocarcinoma patients with progression or secondary clinical resistance to EGFR-targeted TKIs [19]. The last assessed aspect of the procedure was the choice of chemistry and technology. As p.Thr790Met is the major but not the only mechanism of TKI resistance, liquid biopsy should not be limited to the screening of a single mutation in future. Furthermore, recent publications indicate that the existence of p.Thr790Met-neighboring rs1050171 reduces the sensitivity of the ARMS-based p.Thr790Met mutation detection assay and produces a 14.3% false-negative rate [20]. Therefore, it is important to use a CE-IVD test for ctEGFR detection, which has been validated to account for the p.Thr790Met SNP mutation (Q787Q). Not only rapid real-time PCR methods [11], but also MLPA [21], ddPCR and next generation sequencing [1] are used for ctEGFR somatic mutation screening. While preparation of NGS libraries and high-throughput single-sample sequencing seems to be more interesting from a scientific point of view, it is quicker and cheaper to use the qPCR or ddPCR methods.
Conclusions
Liquid biopsy offers new possibilities for inoperable patients and for monitoring targeted therapies in NSCLC. Studies in larger homogeneous groups of patients receiving TKIs should be conducted to evaluate the potential for feasibility of somatic mutation screening in ctDNA for residual disease detection and to determine a complete or major molecular response to targeted treatment.







