Introduction
Hepatocellular carcinoma (HCC) is a global problem, being the sixth most frequent cancer and the fourth major cause of cancer mortality [1]. Hepatitis C virus (HCV)-related liver cirrhosis is thought to be responsible for 80% of HCC cases. Despite years of intensive investigation, the precise molecular pathways driving virus-associated hepatocarcinogenesis remain unclear, and little progress has been achieved in terms of improving diagnosis and prognosis [2]. Searching for alternatives for early identification of HCC will thus help patients with this fatal disease have a better chance of surviving. Alpha-fetoprotein (AFP) measurement is now employed as an additional diagnostic test. Its clinical value is restricted due to its low sensitivity and specificity. As a result, innovative and reliable biomarkers that can enhance the overall clinical care of HCC are urgently needed [3].
According to recent research, the accumulation of epigenetic and genetic modifications may play a role in various phases of liver carcinogenesis [4]. Cancer is associated with abnormal hypermethylation of CpG islands, a major epigenetic regulatory mechanism [5]. Methylated DNA markers (MDMs) are DNA methylation abnormalities reported in a variety of malignancies, including HCC [6].
RAS association domain family protein 1 transcript A (RASSF1A) and cyclin-dependent kinase inhibitor 2A (CDKN2A) are two cell cycle regulator genes that have previously been shown to be deactivated by hypermethylation in malignant tumors [7].
The RASSF1A gene, which is found on the cytogenetic band Chr 3: p21.31, is a key component of the RAS signaling system [8]. Cell cycle regulation, apoptosis induction, microtubule stability, and cellular adhesion are all functions of the protein encoded by RASSF1A [9]. According to previous research, RASSF1A deactivation by promoter hypermethylation has been found to occur frequently in malignant tumors, including HCC [7].
CDKN2A is a well-known tumor suppressor gene that produces the p16-INK4a protein, a cell cycle negative regulator. It inhibits cyclin-dependent kinases 4 and 6 (CDK4, CDK6), which play a key role in cell cycle control by stopping cell progression from the G1 to the S phase, and so assist in cancer prevention [10]. CDKN2A is shown to be downregulated in HCV-induced HCC due to widespread CpG methylation [11]. RASSF1A and CDKN2A methylation frequency in HCC and cirrhotic liver tissue samples has been widely examined [12].
The aim of the present work was to evaluate the methylation status of both genes in the peripheral blood and compare them to AFP and evaluate their relationship with the clinicopathological features of the tumor as potential non-invasive biomarkers for HCC development in Egyptian patients with HCV-related liver cirrhosis.
Material and methods
Study design
The current work was conducted at the Medical Research Institute Hospital, Alexandria University, between December 2020 and June 2021 as a case-control study. All patients were gathered sequentially from the hepatology unit and cancer management and research unit, and all participants provided informed consent. The study was authorized by the Local Ethics Committee of the Medical Research Institute Hospital, which adheres to the Helsinki Declaration’s requirements.
Study population
Thirty healthy volunteers (control group) in addition to forty patients with HCC on top of HCV-associated liver cirrhosis (tumor group), and forty patients with HCV-associated liver cirrhosis (cirrhosis group) participated in this study.
The American Association for the Study of Liver Disease (AASLD) guidelines were used to diagnose HCC, which were based on the presence of an ultrasonic focal lesion and then confirmed with AFP elevation and imaging (contrast-enhanced triphasic computed tomography scan study or magnetic resonance imaging) and/or pathological examination [13]. The staging of HCC was assessed according to the Barcelona Clinic Liver Cancer (BCLC) system [14]. Clinical, biochemical, and ultrasonographic evidence was used to diagnose cirrhosis. The severity of the liver illness was determined using the Child-Turcotte-Pugh (CTP) score and class [15]. Infection with the hepatitis B virus, any cause of chronic hepatitis, and any other malignancy were all ruled out. Antiviral therapy-treated patients were also excluded. The control group consisted of healthy individuals based on the clinical examination and normal laboratory results. They were matched to the included patients in terms of age and sex.
Laboratory investigations
The molecular study and the standard laboratory testing, which included complete blood count, liver function tests, and AFP, were performed after blood sample collection. As part of the routine clinical workup, all patients were tested for viral hepatitis markers such as hepatitis B surface antigen and core antibodies, as well as anti-HCV antibodies.
Molecular study of DNA methylation status
The methylation status of the RASSF1A and CDKN2A genes was determined using methylation-specific polymerase chain reaction (PCR) (MSP).
DNA extraction
The Gene JET Genomic DNA Purification Kit (Thermo Fisher Scientific) was used to extract genomic DNA from whole blood that has been anticoagulated with ethylenediaminetetraacetic acid (EDTA). The purity and concentration of the extracted DNA samples were determined using agarose gel electrophoresis and a Thermo Scientific Nano Drop 1000 Spectrophotometer (Thermo Fisher Scientific, USA). The isolated DNA samples were then kept at –80°C until further investigation.
Bisulfite modification
The isolated DNA was bisulfite modified according to the manufacturer’s instructions using the EpiTect Fast DNA Bisulfite Conversion Kit (Cat No. 59824; Qiagen). Unmethylated cytosine residues in DNA were converted to uracil by bisulfite treatment, but methylated cytosine residues remained intact.
Amplification by PCR
The modified DNA was then amplified using COSMO ‘Hot Start’ PCR RED Master Mix (Willowfort, UK) and the primers provided by Invitrogen, Thermo Fisher Scientific, according to the manufacturer’s instructions. Two MSP reactions were carried out for each gene, one for the methylated DNA sequences and the other for the unmethylated ones using the primers and the MSP conditions illustrated in Table 1. Each MSP reaction was performed in a total volume of 25 µl using a Quanta Biotech, UK thermal cycler. For testing the specificity of the primers, fully methylated and unmethylated bisulfite converted DNA controls (10 ng/µl) (Cat No. 59695, EpiTect PCR Control DNA Set, Qiagen) were included in each MSP run.
Table 1
Primer sequences and PCR thermal cycler conditions of unmethylated and methylated promoter regions of RASSF1A and CDKN2A genes
DNA detection
The resultant PCR products were then separated by agarose gel electrophoresis (2% for methylated and unmethylated reactions of RASSF1A gene, and 2.5% for methylated and unmethylated reactions of CDKN2A gene) and visualized by ethidium bromide staining. The primer sequences and PCR thermal cycling conditions of promoter regions of RASSF1A and CDKN2A genes are presented in Table 1. The specific bands of each reaction are illustrated in Figures 1 and 2.
Fig. 1
Methylated (A) and unmethylated (B) runs of RASSF1A gene on 2% agarose gel. Lane 1: 100 bp ladder. Lane 2: Bands of positive control at 94 bp in the methylated run (A) and at 108 bp in the unmethylated run (B). Lane 3: No bands of negative control (nuclease-free water) in both runs (A) and (B). Lane 4: Unmethylated case with a band in the unmethylated run only. Lane 5: A methylated case with a band in the methylated run only. Lanes 6: A methylated case with bands in both methylated and unmethylated runs
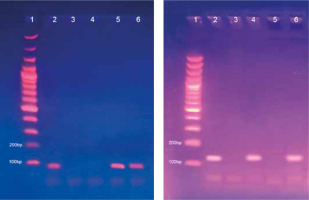
Fig. 2
Methylated (A) and unmethylated (B) runs of CDKN2A gene on 2.5% agarose gel. Lane 1: 100 bp ladder. Lane 2: Bands of positive control at 150 bp in the methylated run (A) and at 151 bp in the unmethylated run (B). Lane 3: No bands of negative control (nuclease-free water) in both runs (A) and (B). Lane 4: A methylated case with bands in both methylated and unmethylated runs. Lane 5: A methylated case with a band in the methylated run only. Lane 6: Unmethylated case with a band in the unmethylated run only
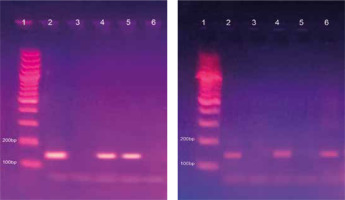
The samples that were amplified from the primer pair for the methylated DNA sequence were considered methylation-positive, whereas the samples that were amplified from the primer pair for the unmethylated sequence were considered methylation-negative. Samples with PCR products from both primer pairs were considered partially methylated, which also represents a methylation-positive status.
Statistical analysis
The data were analyzed using IBM SPSS software package version 20.0 (IBM Corporation, Armonk, NY). To confirm that the distribution of variables was normal, the Kolmogorov test was utilized. For categorical variables, the χ2 test (Fisher’s exact or Monte Carlo correction) was employed to compare groups. The Student t-test was used to compare two groups with normally distributed quantitative variables. ANOVA was used to compare three groups. The Mann-Whitney test was used to compare two groups and the Kruskal-Wallis test was used to compare several groups for non-normally distributed quantitative variables. Receiver operating characteristic (ROC) curve analysis was used to assess the diagnostic performance, and the area under the curve was calculated. A 5% threshold of significance was adopted.
Results
The current study included 40 HCC patients (70% male and 30% female) with a median age of 59.5 years, 40 HCV-associated liver cirrhosis patients (21 males and 19 females) with a median age of 55.5 years, and 30 healthy volunteers (14 males and 16 females) with a median age of 57.5 years.
Table 2 compares the methylation status of the RASSF1A and CDKN2A genes, as well as the levels of AFP, across the examined groups.
Table 2
Methylation status of RASSF1A and CDKN2A genes and AFP level among the studied groups
Univariate regression analysis of RASSF1A gene methylation showed that the odds ratio for predicting HCC was significantly high in the control and cirrhosis groups (4.45 and 2.51 respectively). Moreover, CDKN2A gene methylation had a significantly high odds ratio for predicting HCC in the control and the cirrhosis groups (8.31 and 2.54 respectively) (Table 3).
Table 3
Odds ratio of RASSF1A and CDKN2A gene methylation for predicting HCC in the studied groups
| Gene | Tumor vs. cirrhosis® | Tumor vs. control® | ||||
|---|---|---|---|---|---|---|
| p | OR | 95% CI | p | OR | 95% CI | |
| RASSF1A | 0.045* | 2.51 | 1.019-6.198 | 0.005* | 4.45 | 1.551-12.741 |
| CDKN2A | 0.045* | 2.54 | 1.023-6.298 | < 0.001* | 8.31 | 2.731-25.276 |
The relationship between the methylation status of the RASSF1A and CDKN2A genes and clinical, radiological and laboratory parameters is shown in Table 4.
Table 4
Association between RASSF1A and CDKN2A gene methylation and the clinical, radiological and laboratory parameters of the tumor group (n = 40)
[i] ALT – alanine aminotransferase, AST – aspartate aminotransferase, PT – prothrombin time, INR – international normalized ratio, AFP – α-fetoprotein, BCLC – Barcelona Clinic Liver Cancer, FL – focal lesion, MC – Monte Carlo, FE – Fisher exact, p – p-value for comparing between the different parameters, * statistically significant at p ≤ 0.05
There was a significant association of the methylation status of the RASSF1A gene with the tumor stage, the number of focal lesions, liver lobe involvement, and lymph node metastasis, whereas CDKN2A gene methylation did not show any significant association with tumor characteristics. Regarding the laboratory parameters, RASSF1A gene methylation was significantly associated with increased ALT, AST, PT, and decreased albumin, while CDKN2A gene methylation was significantly associated with increased ALT, AST, and total bilirubin.
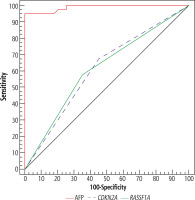
The diagnostic performance of RASSF1A and CDKN2A gene methylation was lower than that of AFP in discriminating HCC patients from cirrhotic patients. They showed moderate sensitivity (57.5% and 67.5% respectively), and moderate specificity (65.0% and 55.0% respectively). The area under the curve (AUC) was 0.613 and 0.0613 respectively. Meanwhile, AFP showed higher sensitivity and specificity (95.0% and 85.0% respectively), with an AUC of 0.989 at a cutoff of 20 ng/ml (Table 5, Fig. 3).
Fig. 3
ROC curve for α-fetoprotein (AFP), RASSF1A, and CDKN2A gene methylation to discriminate HCC patients (n = 40) from cirrhosis patients (n = 40)
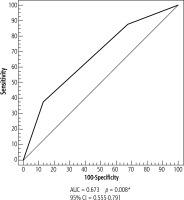
The diagnostic performance significantly improved after combining RASSF1A and CDKN2A gene methylation, to a sensitivity of 87.5%, but with a low specificity of 32.5%, while AUC became 0.673 (Table 5, Fig. 4).
Table 5
Diagnostic performance of RASSF1A and CDKN2A gene methylation versus AFP in predicting HCC in cirrhosis patients
Fig. 4
ROC curve for combined CDKN2A and RASSF1A gene methylation to discriminate HCC patients (n = 40) from cirrhosis patients (n = 40)
Furthermore, RASSF1A gene methylation performed better than CDKN2A gene methylation in distinguishing early (A+B) and late (C+D) BCLC stages of HCC. It had a positive predictive value (PPV) of 82.61%, negative predictive value (NPV) of 52.94%, and accuracy of 70.0% (Table 6).
Table 6
Diagnostic performance of RASSF1A and CDKN2A gene methylation in discriminating early and late BCLC stages of HCC
Discussion
Numerous DNA-methylation-dependent epigenetic processes have been identified in HCC. The tumorigenic process in cancer cells is aided by the transcriptional silencing of tumor-suppressor genes by CpG island promoter hypermethylation [5].
The relationship between methylation of the RASSF1A and CDKN2A genes and carcinogenesis has previously been investigated using various detection techniques, with the overall result being that dysregulation of both genes is linked to tumor growth in HCC [1, 16]. The MSP approach was utilized in this research to evaluate the methylation status of both genes in the peripheral blood as possible non-invasive indicators for HCC in Egyptian patients with HCV-associated liver cirrhosis.
The RASSF1A gene was found to be methylated in 57.5% of the HCC patients included in this study. When compared to the cirrhosis and control groups, it was significantly greater in the tumor group. Previous studies have established that the methylation status of the RASSF1A gene may have a role in the process of hepato-carcinogenesis and may be employed as non-invasive diagnostic biomarkers for HCC, which is consistent with our findings [5, 17]. Furthermore, Xu et al. in their meta-analysis found that aberrant methylation of the RASSF1A gene was significantly correlated with the risk of HCC, and it is a promising biomarker for the diagnosis of HCC from tissue and peripheral blood [1]. On the other hand, Wu et al. revealed no methylated RASSF1A gene levels in the plasma of HCC patients, despite the presence of the hypermethylated RASSF1A gene in the tumor samples [18]. Differences in the detection technique and primers used might explain some of these discrepancies.
In normal hepatocytes, the RASSF1A-encoded protein suppresses cyclin D1 and escapes G1 phase arrest; it has a role in cell cycle regulation and proliferation. Both in HCC cell lines and patients’ liver tissues from hepatitis, cirrhosis, and HCC, an inverse relationship between RASSF1A methylation and its RNA expression has been demonstrated [19]. Because RASSF1A methylation inhibits its tumor-suppressing function, it permits the injured hepatocyte to continue further into the cell cycle, culminating in the growth of a tumor [17].
In the current study, the RASSF1A gene was significantly methylated in advanced tumor characteristics such as the advanced stage, multiple focal lesions, lymph node metastasis, and involvement of both liver lobes. Similar results were obtained in the Hu et al. study where the expression of the RASSF1A gene was linked to the TNM stages of HCC and lymph node metastases and was shown to correlate with the levels of AFP, portal vein thrombosis and capsular infiltration. They postulated that reduced RASSF1A expression was linked to an increased risk of tumor development, metastasis, and recurrence [20].
Our findings demonstrated no significant association between serum AFP levels and RASSF1A methylation status, which is consistent with the findings of Yeo et al., who did not detect a correlation between serum AFP levels and serum RASSF1A gene methylation status [21]. Furthermore, Hu et al. reported that serum RASSF1A methylation status did not affect the biochemical markers including AFP [20]. This contradicts the findings of Zhang et al., who observed a strong association between tissue RASSF1A gene methylation and blood AFP levels in HCC patients [22]. This difference could be due to differences in the study design and cut-off value of AFP.
As a tumor suppressor gene, CDKN2A plays a role in cell cycle regulation and its methylation may prevent transcriptional processes, resulting in gene silence and reduced expression, influencing the cellular biological activities [23].
CDKN2A was methylated in 67.5% of HCC patients in the current work, with a significant difference between the tumor, cirrhosis, and control groups. CDKN2A methylation was identified by Csepregi et al., using MethyLight, and it was found frequently in both non-HCV and HCV-induced HCC samples [19]. In addition, previous research by Su et al. found that CDKN2A gene methylation increased from cirrhotic tissue to HCC [24], which is consistent with our findings that CDKN2A gene methylation was significantly higher in the cirrhosis group than in the control group.
In the present study, no association between CDKN2A methylation and tumor characteristics was detected. This is in line with the findings of Lou et al., who found no relation between tumor number and diameter and CDKN2A methylation [25].
As regards AFP serum levels, there was no significant association with CDKN2A methylation, in contrast to Seif et al., who observed that AFP serum levels were considerably higher in the methylated group of patients than in the non-methylated group [7]. The current study’s findings might be related to the diversity of the patient groups evaluated.
Methylation of the RASSF1A gene was linked to encephalopathy, Child class, and portal hypertension in the current study. It was also linked to higher levels of ALT, AST, and PT, as well as lower levels of albumin. Methylation of the CDKN2A gene was linked to higher levels of ALT, AST and total bilirubin. This suggests that methylation of the RASSF1A and CDKN2A genes may indicate the severity of the underlying HCV-related liver cirrhosis in HCC patients. This might be due to long-term HCV-induced inflammation. Previous studies have revealed that HCV has a direct role in epigenetic regulation. HCV upregulates the methylation state of the RASSF1A promoter through control of SET and MYND domain-containing protein 3 [26]. Furthermore, Park et al. found that the HCV core protein reduces the levels of CDKN2A, via promoter hypermethylation, giving hepatocytes an edge in cell proliferation [27].
We found that methylation of the RASSF1A and CDKN2A genes had a weaker diagnostic performance than AFP in discriminating HCC patients from cirrhotic patients with lower sensitivity and specificity compared to AFP at a threshold of 20 ng/ml. However, the diagnostic performance significantly improved after combining RASSF1A and CDKN2A gene methylation, with higher sensitivity but still low specificity.
The meta-analysis of Zhou et al. reported similar sensitivity for CDKN2A gene methylation, though they observed a higher AUC (0.87) and high specificity (0.93) in HCC blood samples [28]. They argued that the poor sensitivity of the CDKN2A gene would limit its diagnostic use; however, when Zhang et al. combined the three methylation biomarkers CDKN2A, RASSF1A, and P15, the sensitivity increased to 0.84, which was in agreement with our results [29]. Furthermore, Dong et al. observed lower sensitivity but greater specificity for AFP, moderate sensitivity for serum RASSF1A methylation and a specificity of 89.8%. When used together, the sensitivity and specificity of RASSF1A methylation and serum AFP are improved to 80.9% and 93.4%, respectively [30]. These discrepancies in the diagnostic performance could be attributed to the detection methods or sample size, and so its non-invasive application value for HCC needs to be verified and further confirmed.
Furthermore, RASSF1A gene methylation performed better than CDKN2A gene methylation in distinguishing the early and late stages of HCC. To our knowledge, no other studies have used both markers to distinguish between the early and late stages of HCC, but the results need more validation.
In conclusion, the significant methylation of the RASSF1A and CDKN2A genes in the HCC group may reveal their role in hepatocarcinogenesis. Moreover, the combination of both genes’ methylation in the blood could be utilized as a complementary biomarker in the early detection of HCC among high-risk patients with HCV cirrhosis. RASSF1A gene methylation may be considered in distinguishing the early and late stages of HCC.
Our study’s limitations include no estimation of RASSF1A and CDKN2A as prognostic factors in HCC, and no correlation with patients’ survival or response to treatment. We also did not link both genes’ methylation status in blood with that in tumor tissue.
In future prospective investigations on a greater number of patients, the methylation levels of both genes in HCC should be studied further using alternative methodologies.






