Introduction
Humans generally have thousands of microbes and bacteria in their digestive system, which are collectively referred to as the microbiome. The microbiome is the genetic material of all of the microorganisms that live inside the human body, which include, but are not limited to, bacteria and viruses. The bacteria in the microbiome break down toxins, manufacture amino acids, and form barriers against invading viruses [2]. The microbiome plays a vital role in the human body, and without it, it would be very difficult to survive, since they help control the digestion of food, the immune system, and the central nervous system [3]. Even though many types of bacteria and other microbes in your intestines benefit your health, there are some that are unhealthy for you and can lead to diseases. Autoimmune diseases such as diabetes and multiple sclerosis are associated with dysfunction in the microbiome [4]. An imbalance of healthy and unhealthy microbes occurs in some people, called gut dysbiosis. It typically occurs when the bacteria in your gastrointestinal tract become unbalanced. Due to its effects on the human body, such as immune and gut barrier dysfunction, it influences the expression of metabolic diseases and cancers such as inflammatory bowel disease (IBD) and colorectal cancer (CRC) [5]. The idea of the microbiome having such an impact on the human gut will be useful as a reference in future investigations and studies that wish to correlate bacterial make-up with specific diseases.
While there is an association between certain diseases and IBD, a previous study showed that the majority of patients who were being tested on were cured of their IBD once they received secondary to cytotoxic chemotherapy [6]. While the minority of the patients who experienced a flare during their cancer treatment were the ones who received hormonal therapies, for instance, combination cytotoxic chemotherapy with adjuvant hormone therapy, the experiment showed that hormonal therapies for cancer may actually increase the risk of IBD reactivation. This conveys the idea that there could be some sort of link between IBD and certain cancers in patients. This along with the idea that the composition of gut dysbiosis and the developments of tumors could be related to the development of Crohn’s disease or ulcerative colitis helped create a hypothesis [7]. The hypothesis suggests that gut microbiome analysis could be a biomarker for cancer progression in patients who have gastric cancer or colorectal cancer. There could possibly be a link between an imbalance of gut microbes and the development of cancer that is caused by IBD. The study begins with observing the fecal matter that is utilized in order to gather more information on the cancers and identify whether they are progressing due to the imbalance of microbes in the guts or not.
Materials and methods
Two datasets were utilized, the first related to placenta-specific 8 (PLAC8), a type of protein that is encoded by the PLAC8 gene, and its contribution to CRC progression [8], while the second dataset concerns an amplicon sequence analysis of fecal samples from healthy subjects and patients diagnosed with ulcerative colitis or Crohn’s disease [9]. For the first dataset it was determined whether or not gut microbiome analysis could be a predictive marker for cancer patients. For the second dataset, rRNA amplicon analysis was conducted through fecal samples in order to measure the progression of IBD in patients. The first experiment included 19 patients and it was conducted at the Cathay General Hospital. As for the second experiment, approximately 227 patients were taken into account, with them being registered at the People’s Hospital of Hangzhou Medical College. The first step for the research is to gather as much information about the different types of diseases that are being analyzed in this study. Once enough information was collected, Ubuntu and R Studio were used to collect and store the data that were gathered from the two experiments. Ubuntu is a coding platform used for its versatility and extensive developer libraries. It is an open-source platform which let me look into various parts of Ubuntu and source code to understand how it is designed and developed. This software operating system allowed me to store data in files called Sequence Read Archive (SRA) files, which could then be converted into fastq files, objects that make it simple for information to be analyzed in other coding programs. R Studio is used to create visualizations and manipulate data sets. Fastq files were obtained and inputted into R Studio. With this, massive amounts of qualitative coding were categorized, and the information was assigned into groups. This made it easier to facilitate data conversion and measurement comparisons. R Studio was then used to merge read pairings. Taxonomy Analysis could also be utilized in this experiment in order to view certain structures in materials and cells.
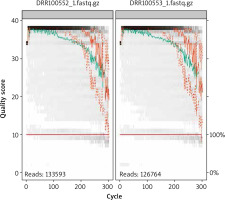
When using gut metabolome analysis, taxonomy analysis was performed. In this taxonomy analysis, the SRA Taxonomy Analysis Tool (STAT) was utilized. STAT calculates the taxonomic distribution of reads from next generation sequencing runs. This exhibits the distribution of reads mapping to specific taxonomy nodes as a percentage of total reads. This makes it easier to visualize the data given and understand it better. STAT map sequencing reads to a taxonomic hierarchy using a two-step plan that identifies organisms matching a read set. Next the organism-specific slices are applied to calculate distribution of the read between taxonomy classes. This tool was used in order to find the difference in percentages in gastric cancer patients compared to the percentage of total reads in healthy subjects. If there is a clear difference between the two, it could one day serve as a biomarker for certain cancers. This is because the dysbiosis of the gut microbiome is associated with host health conditions, and many diseases have shown a correlation with imbalanced microbiota, including IBD and cancer [10]. With this the first experiment was fully analyzed, but it was still unsure whether or not gut microbiome analysis could be a good biomarker for gastric cancer patients. By using the taxonomy to see the percentages in each subject’s total reads, it can be inferred that gut microbiome analysis is a biomarker for certain cancers. This is because the patients who were diagnosed with gastric cancer typically had a higher guanine to cytosine (GC) percentage content. However, they have a smaller percentage of anaerobic bacteria such as Lachnospiraceae or Enterococcus faecium. In genetics, guanine to cytosine content (GC-content) is the percentage of nitrogenous bases in DNA or RNA molecules that are either guanine or cytosine. In this research, dozens of patients were looked at to see whether or not this was the case for the majority of them, which it was, meaning that these two factors could possibly be biomarkers for the progression of cancer caused by IBD. The link may justify and support my hypothesis in which there is a clear progression and characteristics in cancer patients’ fecal samples, that is unlike healthy subjects’ fecal samples. Another method that was implemented in this study was utilizing the coding program Ubuntu [11]. It is an operating system that runs on the desktop or cloud of a personal computer, and it allows for the user to use commands in order to work on datasets and local files. With this, the datasets in this study are converted into SRA files (Fig. 1). Sequence Read Archive files are bioinformatics database files that provide DNA sequencing data, making it easier to process and store all of the information from the datasets. Next R programming, a language and environment for statistical computing and graphics, was used in order to take the SRA files from Ubuntu and convert them into fastq.gz files. Fastq files are used for storing DNA sequencing data and its quality scores [12]. So, R programming will then be able to draw out and create reads that show the quality score and the cycle of them. The next step is to trim the ends of the reads so you can prevent misleading data from being inside the sequencing fragments. Afterwards, data amplicon sequencing data could be used; it will enable analysis of genetic variation in genomic regions. After reading and examining the genetic variation, visualization data could be created in R programming with it, whether it be a table or plots. After downloading the Dada2 package, a set of demultiplexed fastq files corresponding to the samples in your study, you can start inspecting read quality profiles and then calculate the error percentage of the data. After this step, reads will be merged together to obtain full denoised sequences. By doing this, it will be possible to see similarities as well as the differences between patients with IBD and healthy subjects.
Results
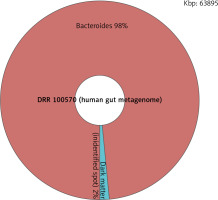
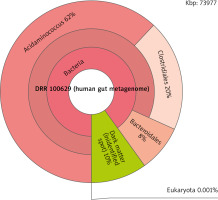
With the taxonomy analysis, it was found that the majority of patients diagnosed with IBD would have a GC-content percentage of 55% or higher while healthy subjects would have an average percentage of 50–53%. This indicates a higher melting temperature than the average person’s. This big difference in melting temperature is not good for polymerase chain reaction (PCR), a technique in molecular genetics that permits the analysis of any short sequence of DNA [13]. It is used to make copies of segments of DNA [14]. Furthermore, the taxonomy analysis enabled the scanning of the human gut metagenome of both the healthy subjects and diagnosed patients. The data showed that the patients diagnosed with CRC or gastric cancer had a higher percentage of bacteria in the gut microbiome analysis than did healthy subjects. The IBD patients generally had a dark matter percentage of 10% or more, while healthy subjects would have 2% or less (Fig. 2, 3). This huge difference in the amount of dark matter in an individual’s human gut metagenome could be an indicator of someone potentially developing a disease.
Fig. 2
The human gut metagenome of an inflammatory bowel disease patient, who has a dark matter percentage of 10%
As for the fastq files, after analyzing the reads with them the DNA sequencer began to produce poor quality scores at around 250 on the cycle, which indicates that it was no longer going to produce longer sequence runs. With this in mind, the data would be informative and useful up until it reached 250. In order to determine whether or not the data are reliable, it is necessary to calculate the error percentage, which in this case is 0.0508%. This suggests that the data utilized in this experiment are credible.
Discussion
There were found to be differences between IBD patients and healthy subjects, as the former had higher GC-content percentages and had higher percentages of dark matter in their human gut metagenome. A high GC can give you G-runs in primers or products, and too many of those in a run may result in intermolecular quadruplexes forming in the PCR mix before or during amplification [15]. This generates complications during primer design and can result in secondary structures, self-dimer formations, and mismatches. Experiments have also shown that DNA containing a higher GC-content percentage tends to have greater resistance to denaturation, and is more stable.
Patients with a higher GC-content percentage often had smaller percentages of anaerobic bacteria, such as Lachnospiraceae and Enterococcus faecium. Lachnospiraceae belong to the core of gut microbiota, and the help metabolize bile acids in the large intestine. Enterococcus faecium is a member of the Firmicutes, and is sometimes used as a probiotic product. Overall, these anaerobic bacteria help prevent the attachment and invasion of pathogenic bacteria into epithelial cells [16]. Pathogenic bacteria can cause infectious diseases. They disrupt normal functions in the human body, sometimes killing cells and tissues. They can also make toxins that paralyze or destroy cells’ metabolic machinery [17]. This can either cause cancer to form or increase the risk that cancer will form. Therefore, with a smaller percentage of anaerobic bacteria found in patients with IBD, they stand at higher risks of cancer secondary to long-standing intestinal inflammation. There are complications with replicating DNA with PCR; however, by using gut microbiome analysis it could be possible to use human gut metagenomes as measurable substances to indicate the development of cancerous cells. Approximately 5 to 10 percent of IBD patients develop colon cancer after 20 years, and 12 to 20% develop cancer after 30 years [18]. It is important to be able to reduce these percentages by preventing the development of cancerous cells and determining the reason for their growth.
The next steps in the study involve using 16S amplicon sequencing on the merge paired reads as well as creating visualization data with the datasets. By employing these steps, it will become possible to understand and correlate IBD and cancer with one another. However, the coding required for this would be too time-consuming for the current work, which means it would be a project for future investigations. One inherent weakness of this study is that we are coerced to stay inside our homes while we do our studies. This makes it more difficult to experiment like you would at a wet bench, where you would perform a wide variety of experiments that deal with biology or chemistry. Another important limitation is the amount of access to assistive technologies such as screen readers being excluded in the studies. With this being said, in my opinion, there is a possible issue of privacy and security of information because the dependability of online information may not be as strong as traditional methods in research. This causes it to be a more intimate and informal way of gathering data and performing procedures. Nonetheless, it may be a sufficient way. The information obtained from future trials will help us understand the chronic intestinal inflammation in IBD and how it may have a significant influence on aiding patients.
Conclusions
Although there is no solid evidence to prove this, we now have sufficient data to support the argument that there is a possible correlation between IBD and colorectal and/or gastric cancer. This study provides an overview of the experimental studies done using PLAC8 and the human gut metagenome. Most of the results showed that IBD patients generally had higher numbers of dark matter and GC-content. They also had chronic intestinal inflammation, something that colorectal and gastric cancer is the result of. However, there are still questions that remain. More research is needed in order to help prove this hypothesis and come up with a potentially effective method that will aid IBD patients. The upcoming years will be a chance to develop this method in order to help as many people as possible.