Introduction
Head and neck squamous cell carcinoma (HNSCC) is the ninth leading type of cancer in the world by incidence, which causes over 90% of epithelial-origin tumours localised in the upper aerodigestive tract [1]. The major risk factors for the development are environmental carcinogens, excessive alcohol and tobacco consumption, and human papillomavirus (HPV) infections [1] – which are driving an increase in HNSCC incidence among younger, non-smoking patients [1]. HNSCC has a high rate of mortality due to metastasis to the regional lymph nodes [2], the tendency to subsequently relapse, and resistance to therapy [3]. However, it has been proven that undergoing an HPV infection is associated with a better prognosis compared to HPV-negative HNSCC patients [1]. Multiple studies have shown that deregulation of different non-coding RNAs (ncRNAs) has an important impact on HNSCC pathology, and they could be used as specific biomarkers in personalised medicine to improve the treatment [4].
Nuclear paraspeckle assembly transcript (NEAT1) is a recently discovered molecule with a critical role in cancer biology [5]. The NEAT1 gene is located on the 11q13.1 chromosome and belongs to the family of ncRNAs of more than 200 nucleotides in length [5, 6]. Although this molecule does not code a protein, it has a very important structural and regulatory function and is a part of complex machinery involving multiple RNA-binding proteins (RBPs) [7, 8], which act as fundamental regulators of gene expression, probably through retention of different molecules in nuclear structures called paraspeckles [8]. NEAT1 also acts as a molecular sponge and regulates the abundance and availability of miRNAs in the cellular environment [9–11]. In recent years, there have been many reports regarding the activity of NEAT1 as an oncogene that promotes proliferation [11–13], inhibits apoptosis and cell cycle arrest [11], regulates blood-tumour barrier permeability, participates in mesenchymal-epithelial transition leading to metastasis, and tumour sensitivity to chemotherapeutics [14, 15]. In addition, NEAT1 promotes the growth of cancer cells, even under hypoxic conditions, and is overexpressed in a wide spectrum of solid tumours resulting in unfavourable overall survival (OS) [11, 14, 16–29] and downregulated in haematological malignancies [30].
In this study, we have analysed the expression of NEAT1 in HNSCC patients’ samples and in HNSCC cell lines as well as using data taken from The Cancer Genome Atlas project. Furthermore, we investigated the correlations between the levels of NEAT1 and some of its target genes in HNSCC patients to determine the role they play in cancer biology.
Material and methods
HNSCC cell lines and patients’ samples
The HNSCC cell lines: SCC-040 (oral cancer model), SCC-25 (tongue cancer model), FaDu (hypopharyngeal cancer model), CAL27 (tongue cancer model), and DOK (dysplastic oral keratinocyte cells from a tongue as a model of healthy tissue) were used for the study and cultivated as described previously [31]. Patients’ RNA samples, tumour and matched adjacent normal, were taken from a previous study [31]. Expression levels of NEAT1 (family) in cell lines and patients’ samples were measured using lncProfiler qPCR Array Kit (SBI) and SYBR Green 2x Master Mix (Roche) as described previously [32]. All real-time PCR data were analysed by calculating the 2-ΔCT, normalising against the mean of reference genes (18S rRNA, RNU43, GAPDH, LAMIN A/C, U6) from the quantification plate.
TCGA data
TCGA expression data of lncRNA NEAT1, as well as the clinical data, were downloaded from the University of California Santa Cruz, cBioPortal, and the UALCAN databases. The above expression values were presented as RNAseq (pan-cancer normalised log2 [norm_count+1]) and mRNA expression z-scores (RNA Seq V2 RSEM).
Data analysis
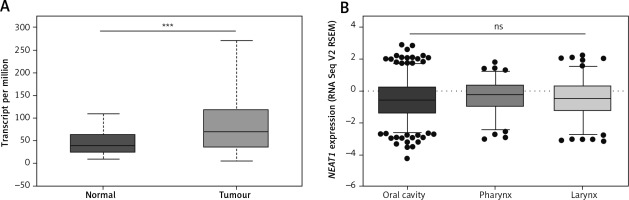
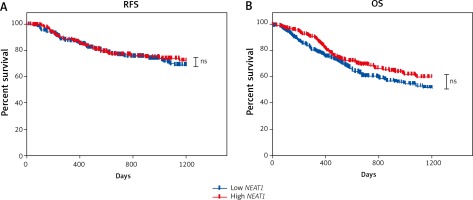
The expression levels of lncRNA NEAT1 were analysed in all HNSCC sample localisations depending on the clinicopathological parameters, such as age (< 61 vs. > 61), gender (female vs. male), alcohol consumption (positive vs. negative), smoking regularly (no/ex vs. yes), cancer stage (I + II vs. III + IV), T-stage (T1 + T2 vs. T3 + T4), N-stage (N0 vs. N1 + N2 + N3), cancer grade (G1 + G2 vs. G3 + G4), perineural invasion (positive vs. negative), lymphoid node neck dissection status (positive vs. negative), HPV p16 status (negative vs. positive), and angiolymphatic invasion (positive vs. negative). The expression level of NEAT1 was also analysed depending on tumour localisation (oral cavity vs. pharynx vs. larynx). The average value of NEAT1 expression was determined in a group of 566 patients, and subgroups were selected based on its high (n = 284) and low (n = 282) expression. Next, relapse-free survival (RFS) and overall survival (OS) were analysed in these subgroups.
Genes correlated with NEAT1
Genes positively and negatively correlated with NEAT1 (Spearman’s correlation > +0.3 or < −0.3, respectively) were obtained from cBioportal (TCGA) and analysed using the REACTOME pathway tool [33].
Functional enrichment analysis and prediction of gene function
Gene set enrichment analysis (GSEA) software version 3.0 was used for the analysis of functional enrichment, as described previously [34, 35]. HNSCC patients were divided into two groups with high and low expression of NEAT1. The input file contained expression data for 20530 genes and 565 patients. The 1000 gene set permutations for the analysis and pathways (the oncogenic signatures [C] and hallmark gene sets [H] and collection from MSigDB) was used, and a nominal p-value ≤ 0.05 and false discovery rate (FDR) ≤ 0.25 were considered as significant. Next, the interactions between protein-encoding genes in the pathway, which were the most significantly enriched in a group of patients with low vs. high NEAT1 expression, were analysed using the GeneMANIA prediction tool [36].
Statistical analysis
All statistical analyses were performed using GraphPad Prism 5 (GraphPad, San Diego, CA,USA) and Statistica 13 (StatSoft Polska). The t-test, Mann-Whitney U test, or one-way ANOVA test were used in analysed subgroups depending on the data normality, which was assessed using the Shapiro-Wilk normality test. In all analyses, p < 0.05 was used as statistically significant. The RFS and OS analyses were carried out using the log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests, respectively.
Results
NEAT1 was up-regulated only in some HNSCC patients’ samples and cell lines
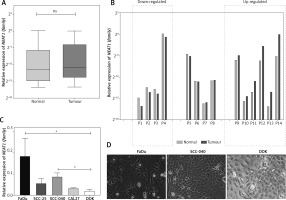
First, the NEAT1 expression was checked in patients’ samples and five HNSCC cell lines. Only in the case of a few patients were upregulation (fold change 2.15–34.73) and downregulation (fold change 0.27–0.48) of NEAT1 (family) observed in tumours, compared to the matched adjacent normal samples (0.1348 ±0.2974 vs. 0.0886 ±0.2910; p = 0.5842) (Figs. 1A and 1B). Moreover, only in the case of two aggressive cell lines, FaDu and SCC-040, was significant up-regulation of NEAT1 (family) expression compared to the dysplastic oral keratinocyte (DOK) cell line observed (p = 0.0154 and p = 0.0479, respectively) (Fig. 1C).
Fig. 1
The expression level of NEAT1 (family) in HNSCC patients’ samples (A and B) and in HNSCC cell lines (C) that display different morphology (D); paired t-test, ns – not statistically significant, * p < 0.05; microscopic pictures of FaDu, SCC-040 and dysplastic oral keratinocyte (DOK) cell lines, 20× magnification
Compared to normal tissues, NEAT1 is up-regulated in most cancers, including HNSCC
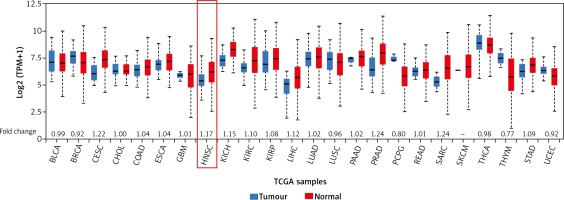
Next, the NEAT1 expression level was checked across 24 different cancers analysed during the TCGA project. In the squamous cell carcinomas, the highest fold change (1.22) of NEAT1 was observed for cervical squamous cell carcinoma (CESC) and the lowest for lung squamous cell carcinoma (LUSC; 0.96); in the case of HNSCC, a 1.17-fold change was indicated. In the group of adenocarcinomas, the greatest fold change of NEAT1 was observed for prostate adenocarcinoma (PRAD; 1.24) and the lowest for rectum adenocarcinoma (READ; 1.01). In the group of other cancers, the greatest fold change of NEAT1 was observed in sarcoma (SARC; 1.24) and the lowest in thymoma (THYM; 0.77). All results are presented in Figure 2.
A significant up-regulation of NEAT1 expression in HNSCC relative to normal samples was observed (68.332 vs. 38.350 transcripts per million; p = 0.0004) (Fig. 3A). Next, based on the National Institute of Health (NIH) classification, patients were divided into three groups according to the localisation of HNSC: oral cavity (n = 346), pharynx (n = 92), and larynx (n = 128). Subsequently, the levels of NEAT1 expression were analysed. No significant differences between tumours in various localisations were observed (p = 0.5058) (Fig. 3B).
Fig. 2
Expression of NEAT1 across 24 TCGA cancers. Graph from UALCAN database, modified; fold change was estimated based on median expression in tumour samples versus normal samples (BLCA – bladder urothelial carcinoma, BRCA – breast invasive carcinoma, CEST – cervical squamous cell carcinoma, CHOL – cholangiocarcinoma, COAD – colon adenocarcinoma, ESCA –esophageal carcinoma, GBM – glioblastoma multiforme, HNSC – head and neck squamous cell carcinoma, KICH – kidney chromophobe, KIRC – kidney renal clear cell carcinoma, KIRP – kidney renal papillary cell carcinoma, LIHC – liver hepatocellular carcinoma, LUAD – lung adenocarcinoma, LUSC – lung squamous cell carcinoma, PAAD – pancreatic adenocarcinoma, PRAD – prostate adenocarcinoma, PCPG – pheochromocytoma and paraganglioma, READ – rectum adenocarcinoma, SARC – sarcoma, SKCM – skin cutaneous melanoma, THCA – thyroid carcinoma, THYM – thymoma, STAD – stomach adenocarcinoma, UCEC – uterine corpus endometrial carcinoma)
The expression levels of NEAT1 differ based on patients’ smoking status
Expression levels of NEAT1 were analysed, determined by group division depending on available clinicopathological parameters in all HNSCC samples. The only significant difference between expression levels of NEAT1 was observed in patients who either smoked regularly or were ex-smokers/non-smokers (p = 0.005), but it is worth noting that there was a considerable difference in the number of patients in these groups (n = 221 vs. n = 20). Other parameters that were analysed did not show any differences between various groups. All data are presented in Table 1.
Table 1
NEAT1 expression levels are dependent on clinicopathological parameters in all localisations of HNSCC
Patients with high NEAT1 expression display slightly better overall survival with close to statistical significance
Next, to determine if lncRNA NEAT1 could be used as a prognostic biomarker, HNSCC samples were divided into low and high NEAT1 expression groups using the mean of NEAT1 expression in all samples as a cut-off. A slightly longer OS of patients with low NEAT1 expression was observed (p = 0.0553). However, there was no difference in RFS time between patients with low and high NEAT1 expression levels (p = 0.6478) (Fig. 4). Moreover, no statistically significant differences in OS and RFS in the case of patients divided into subgroups according to tumour localisation (oral cavity, pharynx, and larynx) were observed (data not shown).
Genes correlated with NEAT1 are involved in mRNA metabolism and cellular transport
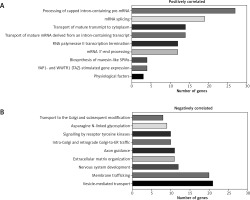
Genes correlated with NEAT1 (Spearman’s correlation > +0.3 or < −0.3, respectively) were analysed, and expressions of 859 were positively (p < 0.05) and 112 negatively (p < 0.05) connected with analysed lncRNA. Analysis of genes positively correlated with NEAT1 indicated that they are involved mostly in mRNA metabolism (transcription, maturation, and transport). For genes negatively correlated, involvement in protein transport and modification as well as membrane trafficking and vesicle-mediated transport was indicated, Figure 5.
Patients with high and low expression of NEAT1 have a different pattern of genes
The functional implications of NEAT1 expression signature were investigated using gene set enrichment analysis (GSEA), and the six top enriched datasets are shown in Figure 6A. It was found that the most up-regulated genes in the NEAT1 low-expressing group of patients were clustered most significantly in the cAMP (the cAMP-dependent pathway), the MYC pathway, the unfolded protein response, the MTORC1 signalling pathway, the oxidative phosphorylation, and DNA repair (NES = 1.745, 1.673, 1.766, 1.470, 1.630, and 1.631, respectively). We identified 66 genes in the cAMP-dependent pathway, 54 genes in MYC pathway, 44 genes in unfolded protein response, 59 genes in the MTORC1 signalling pathway, 91 genes in oxidative phosphorylation, and 67 genes in DNA repair, of which 73.40%, 70.28%, 48.90%, 70.65%, 71.98%, and 54.54% were co-expressed, respectively (Fig. 6B).
Fig. 6
GSEA results of HNSCC patients analysed in groups with low (red)/high (violet) expression of NEAT1. A) GSEA plots of the most enriched datasets, NES (normalised enrichment score), p-value (nominal p-value), FDR q-value (false discovery rate). B) Interactions between protein-encoding genes in the pathways, which were the most enriched in a group of patients with low vs. high expression of NEAT1

Discussion
NEAT1 belongs to the highest regulated lncRNAs among various types of cancer [16]. Moreover, high levels of lncRNA NEAT1 were positively correlated with poor OS [5, 13, 19], cancer stage, and metastasis in the head and neck area, such as in oesophageal and laryngeal squamous cell carcinoma [20, 37]. This molecule can be found in high levels in the cell’s nucleus and cytoplasm [38], where it might induce apoptosis or promote invasion [27], stem cell-like phenotype, epithelial-to-mesenchymal transition (EMT), and resistance to various therapies [14]. Accordingly, NEAT1 might prove a good, prognostic biomarker of HNSCC. Thus, the elucidation of the NEAT1 expression in HNSCC patients’ samples, cell lines, and TCGA data were analysed. Moreover, the NEAT1 network and its target genes, patients’ clinicopathological parameters, and the impact of these interactions on disease pathogenesis data from TCGA databases were used.
The first important finding was that NEAT1 was up-regulated in some HNSCC patients’ samples and in invasive cell lines. However, the TCGA data indicated significant up-regulation of NEAT1 in HNSCC tissue compared to the healthy samples. Moreover, significant differences in the expression levels between cancers localised in the oral cavity, pharynx, or larynx were seen. In contrast to Chen et al., we did not observe the association of high NEAT1 expression with tumour stage [20]. Moreover, we only found a statistically significant difference between the groups of smokers and non-smokers. However, the disproportion in the number of patients in these subgroups was substantial. Information from databases provides the opportunity to analyse a large number of patient phenotypes, but this also makes it impossible to select more evenly distributed subgroups of individuals, which is a limitation of this study. Only the OS analysis of HNSCC patients detected a difference close to statistical significance. Surprisingly, patients with higher expression of NEAT1 tend to present slightly better survival outcomes. This is interesting because our results contradict other reports of the negative impact of high NEAT1 expression levels on patients’ survival [5, 11, 12, 16–18, 20–22, 25–27, 29, 37].
Indeed, in our analysis, we used data for which the NEAT1 isoforms present in the samples were not distinguished. In recent years, there has been growing evidence indicating the need to analyse lncRNA NEAT, not as a whole but considering its two isoforms, NEAT1_1 (3.7kb) and NEAT1_2 (22.7kb), which seem to display completely distinct roles in cancer pathogenesis [8, 39]. Wu et al. proposed that in colorectal cancer, NEAT1_1 might act as an oncogene, promoting cell proliferation, while NEAT1_2 was pivotal for paraspeckle formation [8] and might play a suppressor role [40]. Moreover, comparing the expression of these isoforms in liver metastatic lesions with adjacent normal colorectal tissue and primary colorectal tumour demonstrated significant overexpression of NEAT 1_1 in metastatic tissue [40]. In mice, Nakagawa et al. showed that the expression of lncRNA NEAT1 isoforms is tissue-specific, which might indicate a variable ratio of these molecules in different types of cancers [41]. These changes might occur dynamically, leading NEAT1 to take on oncogene or tumour suppressor functions [39]. Accordingly, the association of high NEAT1 expression with patient survival observed in our study might be due to the greater amount of NEAT1_2 over NEAT1_1. However, we have no direct evidence to validate the finding (hypothesis) because access to the entire TCGA data is restricted. Moreover, in multiple studies of NEAT1 expression in cancer tissues, the issue of its isoforms was either not investigated or not reported [5, 6, 20, 27, 29]. Our analyses highlight the contribution of isoforms of the above lncRNA to cancer pathogenesis and indicate that it is an important aspect that should be further studied.
One of the key factors contributing to the effect of NEAT1 expression levels on tumour development is its interaction with the p53 protein. Interestingly, NEAT1 is a direct transcriptional target of p53, which is a suppressor that is mutated in approximately half of the human cancers [42]. Idogawa et al. analysed the effect of NEAT1 expression level on the prognosis of patients subgroups with and without mutations in the TP53 gene [43]. These authors confirmed their previously formulated thesis, which assumed that NEAT1 supports the suppressor function of the p53 protein. However, this statement seems to apply to wild-type TP53 only, because its mutation alters the function of lncRNA NEAT1, which becomes an oncogene and promotes tumour proliferation. Moreover, in an analysis of the survival probability of patients with 32 types of cancers, 11 of them with high levels of NEAT1 and wild-type TP53 showed longer survival [43]. Moreover, the higher expression of NEAT1 was observed by us in FaDu cell line, which has mutation in TP53 (missense mutation in codon 248), CAL27 (missense mutation in codon 193), SCC-25 and SCC-040 are wild type, but some data indicate mutation in the case of SCC-25 cell line [44, 45]. It should be noted that the frequently used DOK cell line also possesses changed TP53 [46]. In spite of this, data from the UALCAN database indicated no significant differences in NEAT1 expression levels depending on TP53 status in HNSCC. We suspect that TP53 changes the biological function of NEAT1 rather than its expression level. The in vitro analysis of the biological role of NEAT1 depending on different mutations in TP53 should explain this phenomenon in the future.
In mice, Mello et al. indicated that NEAT1 is induced by DNA damage, and its overexpression can suppress transformation in various cell types due to an increase in the number of paraspeckles [47]. The relationship between NEAT1 and TP53 expression is ambiguous for all types of cancers, thus requiring more extensive research.
To better understand the lncRNA NEAT1 interaction network, including its target genes, we checked the gene correlation and we used GSEA. We found that positively correlated genes are connected with mRNA metabolism, and in the case of negatively correlated genes they are connected with cellular transport. Moreover, in patients with low levels of NEAT1, the most enriched genes were clustered in the cAMP-dependent pathway, the MYC pathway, the unfolded protein response, the MTORC1 signalling pathway, oxidative phosphorylation, and DNA repair in a group of patients with low levels of NEAT1. Here, it should be emphasised that we indicate that patients with low NEAT1 expression levels have a worse OS compared to patients with high NEAT1 expression.
The cAMP-dependent pathway is enriched in HNSCC patients with low NEAT1 expression, which is an important signal transduction pathway connecting the internal environment of the cell with external stimuli, such as hormones or cytokines [48]. It has been proven that, with impaired expression, this second messenger has oncogene properties that are responsible for the activation of protein kinase A in selective epithelial tumours [49, 50].
MYC is a proto-oncogene whose expression (under physiological conditions) is strictly controlled by genetic and epigenetic mechanisms [51]. In most cancers, there is pathological activation or overexpression of MYC [52], resulting in cancer growth, inducing stemness, and promoting angiogenesis [51]. It has been demonstrated that following the loss of p53 suppressor function, the MYC protein promotes tumourigenesis [53].
Low expression of lncRNA NEAT1 is also associated with the up-regulation of genes involved in the unfolded protein response. This pathway is responsible for preventing the negative effects of improper protein folding, which acts cytoprotectively not only on healthy cells but also on some cancer cells [54, 55].
The MTORC1 signalling pathway is a complex network involved in the activation of protein synthesis and the promotion of cell growth [56, 57]. Disturbed activation of this pathway in tumours leads to better survival and excessive proliferation [58].
Oxidative phosphorylation (OXPHOS) genes are also overexpressed in a subgroup of patients with low levels of NEAT1. It has been shown that hypoxia present in the tumour of the HNSCC plays a major role in cancer biology and is promoted by OXPHOS up-regulation [59, 60].
Also, patients with low NEAT1 expression had enriched expression of genes involved in DNA repair. The up-regulation of DNA damage response genes leads to resistance to treatment and the development of the ability to metastasise [61, 62].
To summarise, the influence of lncRNA NEAT1 on oncogenic pathways in HNSCC patients supports our observations about the survival rate depending on the NEAT1 expression levels. It seems likely that high expression of NEAT1, through the downregulation of cAMP and MYC pathways, influences the unfolded protein response, the MTORC1 signalling pathway, oxidative phosphorylation, and DNA repair, and thus creates a specific cellular phenotype that is clinically manifested by better patient survival.
Conclusions
The major findings of this study are: (1) NEAT1 expression is up-regulated in some patients’ samples and cell lines. Moreover, the TCGA revealed that NEAT1 is up-regulated in cancer compared to normal tissue in most solid tumours, including HNSCC; (2) Smoking tobacco has a significant impact on NEAT1 expression in patients with HNSCC; (3) Patients with high levels of NEAT1 demonstrate slightly better overall survival with close to statistical significance; (4) Low NEAT1 expression is associated with the up-regulation of oncogenic signalling pathways, such as cAMP, MYC, unfolded protein response, MTORC1, oxidative phosphorylation, and DNA repair.