Introduction
The World Health Organization classification of breast cancer is based on histological criteria [1, 2] such as tumour-node-metastasis score, grading, and hormone receptor expression. Invasive carcinoma of non-special type is the most common type of breast cancer, accounting for 70–80% of all cases. In addition to these criteria, molecular pathological features are increasingly coming upfront. Perou et al. [3] further defined the molecular subtypes of invasive, unspecified breast cancer. The categorization into luminal A and B, Her2-type (HER2)/neu type, and triple-negative or basal breast cancer subtypes is based on gene and receptor expression of the tumour cells. This classification forms the basis of modern chemo- and endocrine therapy, in addition to primary surgical treatment. The oestrogen receptor-expressing luminal subtype is treated anti-hormonally with the selective oestrogen receptor modulator tamoxifen for 5 years, usually after primary surgical therapy. Aromatase inhibitors such as letrozole or anastrozole are favourable for the treatment of postmenopausal women [4, 5]. The adjuvant endocrine therapy has significantly improved the prognosis of breast cancer. However, treatment with tamoxifen can result in acquired resistance against the drug in 20–30% of cases [6]. Additionally, in nearly one-third of all patients, the tumours are already tamoxifen insensitive when therapy starts [7]. Cell culture models are used for in-vitro investigation of the development of tamoxifen resistance [8]. Several mechanisms as well as potential predictive biomarkers for the development of acquired tamoxifen resistance have been identified by these means [9]. In a previous study, we found that the adaption of MCF-7 breast cancer cells to tamoxifen resulted in decreased expression of the long non-coding RNA LINC00992 [10], and we proposed a function in the development of resistance.
Long non-coding RNAs (lncRNA) have a length of more than 200 bases and do not code for a protein. Long-non-coding RNA have mainly regulatory properties and can be classified by their mode of function and genomic localisation [11, 12]. Genomically, lncRNA is localized either within an intron, as long-intronic-non-coding RNA, or between 2 protein-coding genes, as long-intergenic-non-coding RNA (LINC) [12, 13]. LINC00992 is intergenically localized between 2 protein-coding segments.
Long non-coding RNAs have a variety of functions. They can affect gene expression by interacting with specific proteins, mostly transcription factors, as lncRNA-scaffolds or lncRNA-decoys to form ribonucleoprotein complexes that regulate gene expression [14]. These ribonucleo- protein complexes bind to promoter regions to initiate or inhibit gene expression [11]. The lncRNA HOTAIR contributes to carcinogenesis in a variety of tumour enti-ties in this manner [15]. Furthermore, ribonucleoprotein complexes can be transported by a guide lncRNA to specific chromatin loci [16]. Long non-coding RNA can also activate or inhibit enzymes through interactions with these proteins [12]. Also, signalling cascades can be affected by lncRNA [11, 17]. Competing endogenous RNA, or lncRNA-sponges, competes with microRNA for binding sites on mRNA during post-transcriptional regulation thereby fine-tuning miRNA activity. Such fine-tuning is significant for the adaption of cells to stress or changing environmental factors [18–20] such as the adaption of breast cancer to tamoxifen, which is, among other factors, influenced by long non-coding RNA [11].
For breast cancer, several lncRNAs have been discovered that contribute to oncogenesis and the development of tamoxifen resistance. This includes urothelial cancer associated 1 (UCA1), which is highly expressed by tamoxifen- resistant breast cancer cells. These cells showed increased viability and lower apoptosis rates in cell culture [21]. Silencing of lncRNA UCA1 in tamoxifen-resistant cells restored sensitivity to tamoxifen. Conversely, overexpression of UCA1 resulted in resistance against tamoxifen [22]. Long non-coding RNA H19 is also overexpressed by tamoxifen- resistant breast cancer cells. Knockdown of the gene led to sensitivity against tamoxifen [23]. In addition to the development of resistance against tamoxifen, both lncRNAs contributed to the progression of breast cancer [24]. LINC00894-002 expression, such as the expression of LINC00992, is decreased in tamoxifen-resistant MCF-7 cells. LINC00894-002 is decreased via the TGF-β-ZEB1 pathway, which leads to the development of tamoxifen- resistance [25]. Compared to normal tissue, increased expression of LINC00992 is found in breast cancer and is associated with poor prognosis [26].
Long non-coding RNA LINC00992 is part of a regulatory network of mRNA, miRNA, lncRNA, single nucleotide polymorphisms (SNPs), and methylations that influence each other, as well as genes associated with resistance against cytotoxic or endocrine therapeutics in prostate carcinoma [27]. LINC00992 was included into a risk model of 11 autophagy- related lncRNAs as an independent prognostic factor of breast cancer, in which increased expression of LINC00992 is found to be associated with a higher risk [26].
Material and methods
Cell lines and cell culture
MCF-7, T47D, UACC3199, HS578T, MDA-MB468, SkBr3, and MDA-MB-231 cells were obtained from the American Type Culture Collection (ATCC®, Manassas, VA, USA). Tamoxifen-resistant derivatives of MCF-7 cells (MCF-7-TAM-R) were generated from MCF-7 cells by continuous exposure to 4-hydroxytamoxifen, an active metabolite of tamoxifen [10, 28]. All cell lines were cultured in RPMI 1640 medium containing 10% FBS and 1% penicillin/streptomycin but no phenol red. To maintain tamoxifen resistance of MCF-7-TAM-R cells, 10 nM 4OH-tamoxifen was continuously present in the medium [10], corresponding to a dose achieved therapeutically in patients with breast cancer [29]. As a result, 0.01% ethanol was included in the cell culture medium as a solvent. Cells were grown at 37°C and 5% CO2 in a humidified incubator.
Transfection
For transfection of siRNA and plasmid vector, TAM-R and MCF-7 cells were seeded to 30% confluence in a 6-well plate in 1600 µl medium per well. Three wells were transfected with either the plasmid or siRNA; these formed the experimental group. The remaining 3 wells represented the control group and received either a control siRNA or a plasmid without insert. Up-regulation of LINC00992 was performed via transient transfection with the vector pTarget (PN: A1410) by Promega GmbH (Walldorf, Germany), into which the cDNA of LINC00992 was previously integrated using the In-Fusion HD Cloning Plus CE Kit (PN: 638916) by Takara (Kasatsu, Japan). LINC00992 cDNA (3168 bp) was amplified from a plasmid (MGC clone ID 5297581) obtained via Horizon Discovery (Cambridge, UK) (forward primer: AGCATCTCTGCCAACTCTAGAGAGAG, reverse primer: TGTTTTTCCAGTATTTTTATGTAAATTTC AACTATACAGAC). Viromer RED (PN: VR-01LB-01) was used for transient transfection of 2 µg per well of the constructed plasmid vector by BioNTech Delivery Technologies GmbH (Halle [Saale], Germany). Silencing of LINC00992 was performed by transient transfection with DharmaFECT 2 Transfection Reagent (Dharmacon, Lafayette, USA) of a total of 4 siRNA (SMARTpool, PN: R-182242-00-0005, Horizon Discovery, Cambridge, UK) into MCF-7 cells in 3 wells of a 6-well plate. The remaining 3 wells received control siRNA (PN: D-001810-10-05) by Dharmacon (Lafayette, USA). The final concentration of siRNA was 25 nM.
Gene expression analysis
For RNA Purification, the NucleoSpin RNA Mini Kit (PN: REF 740955.50) by Machery-Nagel (Düren, Germany) was used. The cDNA was synthesized using Bioscript reverse transcriptase, oligo dT primers, and random hexamer primers (Bioline Lückenwalde, Germany) according to the manual. Gene expression was monitored using the nCounter® FLEX (NCT-SYST-FLEX) system and nSolver® software (Ver. 4.0) by NanoString Technologies Inc. (Seattle, USA) and a LightCycler® 2.0 instrument controlled by LightCycler® Software 4.1 by Roche Molecular Systems (Basel, Switzerland). Measurement with the nCounter® system was performed as previously described [10] using RNAs from 3 independent experiments each. Pairs were formed with each sample and its corresponding control. Relative expression was calculated from the experimental treatment relative to the control of the same experiment. To quantify relative expression, RPL13 was applied as a housekeeping gene as it was most stable in our earlier experiments [10]. Data were logarithmized to base two, and this log2Fc value used as a measure for expression changes. Quantitative real-time polymerase chain reaction (qRT-PCR) was done as previously reported using RPL13 as a housekeeping gene [10]. Relative expression was calculated by the 2∆CT method [30].
Proliferation, colony-forming, and scratch assay
For the proliferation and viability, colony forming, and scratch assays, MCF-7 and MCF-7-TAM-R cells were transfected in a 6-well plate with the LINC00992 overexpression plasmid (pTarget-LINC00992) and pTarget as control. Cells were counted manually using a Neubauer counting chamber.
For the proliferation assay, 25,000 transfected cells were seeded in 1 mL in each well of a 24-well plate. For 3 consecutive days, a resazurin viability/proliferation test (1 µg/ml) was performed with 6 replicates every 24 hours. Fluorescence was determined after 8-hour incubation at 37°C in a BMG microplate reader with excitation set to 544 nm and emission to 590 nm. A blank was determined by adding resazurin to medium without cells. Fluorescence values were corrected for the blank result, normalized to the result of the first day and to the control group to assess changes in proliferation and viability over time in comparison to control cells.
For colony forming assay, 500 transfected cells were seeded in 3 ml medium into each well of a 6-well plate, including 3 wells each from the experimental group and 3 from the control group. These cultures were incubated for 11 days. On the seventh day a medium change was performed. After this proliferation phase, the cells were washed and fixed with 10% formalin solution and stained with crystal violet. The plates were photographed and analysed using ImageJ according to the scheme of Rafehi et al. [31]. The Analyse Particles function was used to count colonies ranging in size 10–1000 pixels, report area, mean, maxima, minima, standard deviations, and number.
For the scratch assay, transfected cells were seeded in 8 wells of a 24-well plate for each group, and they reached confluence the next day. A mark was placed on the bottom of each well to allow the same spot to be photographed each time. After 24 hours of incubation for adaptation and adherence formation, the medium was replaced with serum-free medium to deprive the cells of their nutrient base to stop further cell divisions. After another day of incubation, a scratch was drawn through the cell lawn using a 200 µl pipette tip (SurPhob tips 200 µl, colorless, Art. No. VT0145, Biozym, Oldendorf, Germany). Subsequently, the medium was replaced by fresh serum-free medium. The marked site was then visited under the microscope and photographed at 10× magnification. Additional photographs were taken after 24 and 48 hours. The images were analysed using ImageJ. For this, 5 strokes per image were drawn orthogonally over the edges of the scratches and their length was measured. The length in number of pixels was converted to µm. The respective length difference of the scratches of the experimental group and control groups of an interval were compared. The length difference was calculated by subtracting the length of one of the 5 strokes of the following day from that of the previous day. Thus, measurement results were obtained for the intervals from the starting point to after 24 hours and from 24 hours to after 48 hours. In addition, the difference in length over 48 hours was compared with the starting point to record the total migration of cells during the experiment.
Database analysis
The expression of LINC00992 in other breast cancer cell lines was examined using cBioportal [32, 33]. For this purpose, the dataset from the Cancer Cell Line Encyclopedia (CCLE) [34] was used. Expression analysis was performed using GEPIA2 [35] to investigate whether LINC00992 are differentially expressed in breast carcinoma subtypes. Survival analysis was also performed to clarify whether high or low expression of LINC00992 is prognostic for disease progression. In addition, LINC00992 expression was analysed in additional tumour entities. GEPIA2 uses TCGA [36] and GTEx [37, 38].
Statistical analysis
The differences of relative expression changes in the nCounter® and qRT-PCR analysis were statistically analysed by using a paired 2-tailed T-test (α = 5%) with Microsoft Excel. A 2-tailed T-test was also used to detect significant differences in the mean area and number of colonies between the control and experimental groups of the colony- forming assay, as well as with the fluorescence values of the proliferation/viability assay and the length difference of the scratch assay. The significance of the expression of LINC00992 in different cell-lines was measured using the ANOVA test with Bonferroni correction.
Results
Expression analysis of LINC00992
We previously established the down-regulation of LINC00992 in MCF-7 upon long-term 4OH-tamoxifen treatment [10]. Here we extended these analyses to other breast cancer cell lines by accessing the CCLE database containing 39 breast carcinoma cell lines via the cBioPortal [32, 33].
In 27 of these, LINC00992 was detected, including MCF-7 (Fig. 1). The cell line T47D showed the highest expression of LINC00992, about 5 times higher than in MCF-7. In all other 25 cell lines, the expression was lower. It was also striking that the highest expression of LINC00992 was found for luminal breast cancer cell lines. For validation, the relative expression of LINC00992 was also measured from RNA samples of different cell lines including T47D and MCF-7 by qRT-PCR (Tab. 1). Here, LINC00992 was also found to be expressed about fivefold higher in T47D than in MCF-7. This experimentally supported the results of the database analysis [10].
Fig. 1
Expression of LINC00992 in various breast cancer cell lines RNA expression
LA – luminal A, LB – luminal B, HER2 – Her2-type, TN – triple-negative Data from the Cancer Cell Line Encyclopedia database were analysed using cBioportal. RNA expression in reads per kilobase million is indicated. Data are sorted in ascending order. T47D has fivefold higher expression of lncRNA than MCF-7. LINC00992 is lower expressed in all other cell lines.
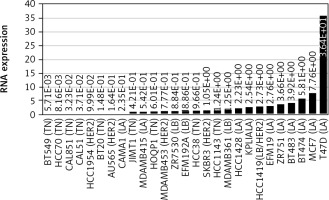
Table 1
Expression of LINC00992 in breast cancer cell lines determined by quantitative reverse transcription polymerase chain reaction
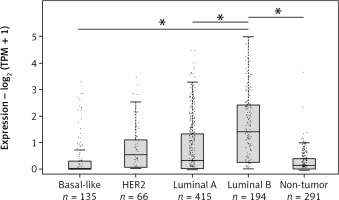
In order to further investigate the expression of LINC00992 in the 4 different intrinsic subtypes by using real breast cancer cases, gene expression analysis was performed with the online database analysis software GEPIA2 [35]. Here, a total of 808 cases from the datasets of the online databases TCGA and GTEx were compared to a control group of 291 samples of typical breast tissues. LINC00992 showed the highest expression in the luminal B subtype, compared to the other molecular subtypes and typical breast tissue. This difference was shown to be significant in a one-way ANOVA assay (p < 0.05). The triple-negative subtype (basal-like) showed the lowest expression of LINC00992 (Fig. 2).
Fig. 2
Expression of LINC00992 in the 4 molecular subtypes of breast cancer expression – log2 (TPM+1)
Analysis of 808 breast cancer cases and 291 control tissues from the TCGA and GTEx databases using GEPIA2. The expression level is plotted as TPM (transcripts per million) + 1 logarithmized to base 2. Breast cancer subtypes are shown in light grey and typical breast tissue in dark grey. LINC00992 is most highly expressed in luminal B type tumours and increased compared with typical breast tissue. These differences were found to be significant in a one-way ANOVA assay by GEPIA2 (p < 0.05). LINC00992 is least expressed in basal-like subtype (triple negative). In the HER2 and luminal A subtypes, the expression level is comparable, with slightly lower expression in luminal A.
Impact of LINC00992 on survival of breast cancer patients
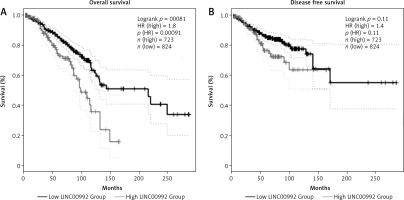
To examine the impact of LINC00992 on the prognosis of breast cancer, survival plots were generated with GEPIA2 using breast cancer cases from the TCGA and GTEx databases. This comprises about 1160 cases. We examined the overall survival (OS) and disease-free survival (DFS) of all breast cancer cases and also for the 4 subtypes. We first optimized the cut-off values for obtaining the best discrimination of all carcinoma cases. The hazard ratio (HR) continuously increased for OS when shifting the cut-off to a lower percentage, and it remained significant from 40/60 (high %/low %) to 10/90. For DFS, however, HR was significant only for the 20/80 and 10/90 (cut-off high %/low %) ratio (Fig. 3 and Suppl. Figs. 1–3). We therefore set the cut-off to the optimized value (23/77) for further subgroup analysis. High expression of LINC00992 is associated with poor prognosis in the OS analysis of all breast cancer cases. Median survival for this group is approximately 100 months (8 years). Cases with low expression of LINC00992 had a better median survival time of approximately 200 months (18 years, HR = 1.8, p = 0.00081). In subgroup-specific analysis, however, only luminal A cases exhibited a significant difference in OS with respect to LINC00992 (HR 2.3, p = 0.0043) (Suppl. Fig. 2). For this group, the LINC00992 low expressing cases had a median OS time of approximately 140 months (11 years) compared to the high expressors, which had a median OS time of almost 100 months (8 years). Analysis of disease-free sur-vival showed no significant differences with this cut-off value for the other molecular subtypes or for all cases (Suppl. Fig. 3).
Fig. 3
Survival analysis comparing LINC00992 high and low expressing cases of all breast carcinoma subtypes overall survival (A), disease-free survival (B)
High cut-off (light grey) was set at 23%, low cut-off (dark grey) at 77%, first standard deviation is represented by dotted lines. The differences in overall survival in both groups was highly significant (p < 0.01) by logrank test. High expression of LINC00992 has a shorter median survival time of 100 months compared to low expression of LINC00992, showing a median survival time of about 220 months (A), no significant differences could be detected for disease-free survival (B).
Effects of modulating LINC00992 abundance in cell lines
Gene expression analysis
To examine a gene-regulatory function of LINC00992, this lncRNA was transiently overexpressed and down- regulated in MCF-7 and MCF-7-TAM-R cells (Suppl. Fig. 4, Suppl. Table 1). Subsequently, the gene expression of these cells was analysed to determine resulting changes. Using siRNA, the relative gene expression of LINC00992 in MCF-7 cells determined by qRT-PCR was decreased by 37% compared to the control group (p < 0.05). Up-regulation of LINC00992 by the plasmid vector led to a nearly 100-fold higher expression, which was nearly significant in a 2-sided T-test for MCF-7 (p = 0.053) as determined by nCounter® analysis (Suppl. Table 2, 3). In MCF-7-TAM-R cells, LINC00992 was only over-expressed to compensate for its decreased expression during Tamoxifen adaption [10].
Fig. 4
Relative expression of ubiquitin D (A), zinc finger protein 462 (B), and single-minded homologue 1 (C)
Gene expression, determined by quantitative real-time polymerase chain reaction, was normalized towards RPL13 and the control treatment. LINC00992 was either down-regulated by siRNA or up-regulated by plasmid-mediated transient over-expression (up). The first standard deviation was given as the error interval. ** p < 0.01, * p < 0.05.
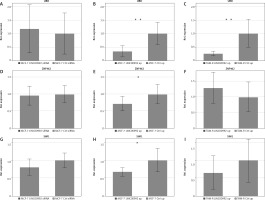
Table 2
Gene expression changes in response to LINC0992 up- and down-regulation in MCF-7 and a tamoxifen-adapted cell line MCF-7-TAM-R determined by quantitative reverse transcription polymerase chain reaction
Table 3
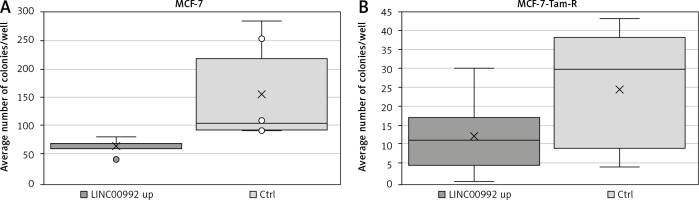
Statistical analysis of colony-forming assay of LINC00992 overexpressing MCF-7 and MCF-7 Tam-R cells
We first determined gene expression by the nCounter® technique with focus on tamoxifen-regulated genes [10] as well as mRNAs included into the PAM50/Prosigna diagnostic panel. Altogether, this included 92 genes and 42 genes associated with the development of tamoxifen resistance [10] and was applied only for a limited number of samples (n = 3) and MCF-7 cells. This experiment represented an initial screen of whether LINC00992 might have any effects on gene expression. The analysis covered 3 samples from MCF-7 cells in which LINC00992 was either down- or up-regulated, as well as 3 control samples each, which received either a control siRNA or the empty plasmid. Data from the nCounter® analysis are shown in Suppl. Table 2, 3. Ubiquitin D (UBD) was especially affected by LINC00992. The gene was increasingly expressed (log2Fc = 0.6) after down-regulation of LINC00992 (p = 0.407), and at the same time the expression of UBD was shown to be decreased (log2Fc = –1.19) after up-regulation of LINC00992 (p = 0.127).
For the other genes, only moderate changes were observed. The expression of ABC-A12 decreased slightly (log2Fc = –0.25) in MCF-7 cells with diminished expression of LINC00992 (p = 0.057). G-protein-coupled oestrogen receptor 1 expression was increased (log2Fc = 0.15) after up-regulation of LINC00992 in MCF-7 (p = 0.087). The expression of matrix metalloproteinase 11 (MMP11) was moderately but significantly decreased (log2Fc = 0.09) after LINC00992 was overexpressed in MCF-7 cells (p = 0.039). We then applied qRT-PCR to further support these data for selected genes using more samples, and we added the oestrogen receptor ESR1 and carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) to the analysis. Data are summarized in Table 2, Figure 4, and Suppl. Figure 5, Tables 2–5. The gene expression analysis by qRT-PCR revealed that the oestrogen receptor alpha coding gene ESR1 was not influenced by expression changes of LINC00992 in MCF-7 or MCF-7-TAM-R cells. The G-protein coupled oestrogen receptor 1 showed a trend toward decreased expression (log2Fc = –0.77) in MCF-7 after down-regulation of LINC00992 (p = 0.062) by qRT-PCR measurement, corresponding to its increased expression after up-regulation detected by nCounter®. However, by qRT-PCR no significant changes could be detected upon up-regulation in MCF-7 and MCF-7-TAM-R. The expression of UBD was again reduced after up-regulation of LINC00992 in MCF-7 (log2Fc = –0.35) and MCF-7-TAM-R (log2Fc = –0.25). Both gene expression changes were highly significant in the 2-sided t-test (MCF-7: p < 0.01, MCF-7-TAM-R: p < 0.01). No change was seen due to down-regulation of LINC00992 in MCF-7 (Fig. 4). The genes single-minded homologue 1 (SIM1) and zinc finger protein 462 (ZNF462) both showed significantly decreased expression following the up-regulation of LINC00992 in MCF-7 cells (SIM1: log2Fc = –0.68, p < 0.05; ZNF462: log2Fc = –0.72, p < 0.05). There were no gene expression changes of both genes after up-regulation of LINC00992 in MCF-7-TAM-R and under decreased expression of LINC00992 in MCF-7 (Fig. 5, 6). The genes of the membrane adhesion molecules CEACAM5 and CEACAM6 showed a trend towards increased expression in MCF-7-TAM-R cells after up-regulation of LINC00992 (CEACAM5: log2Fc = 0.74, p = 0.055; CEACAM6: log2Fc = 0.45, p = 0.130). In MCF-7 cells, these genes were not affected by either up- or down-regulation of LINC00992 (Suppl. Fig. 5, Table 6).
Fig. 5
Proliferation and viability of LINC00992 overexpressing MCF-7 and tamoxifen-adapted MCF-7 breast cancer cells
Results of the resazurin viability assay for MCF-7 cells experimentally overexpressing LINC00992 by a plasmid vector (LINC00992 up) and a corresponding control group receiving the plasmid without insert. Proliferation and viability were inferred from the fluorescence values of resorufin and normalized to the first day and the respective control group. The experiment was repeated 3 times with 8 replicas each. ** p < 0.01.
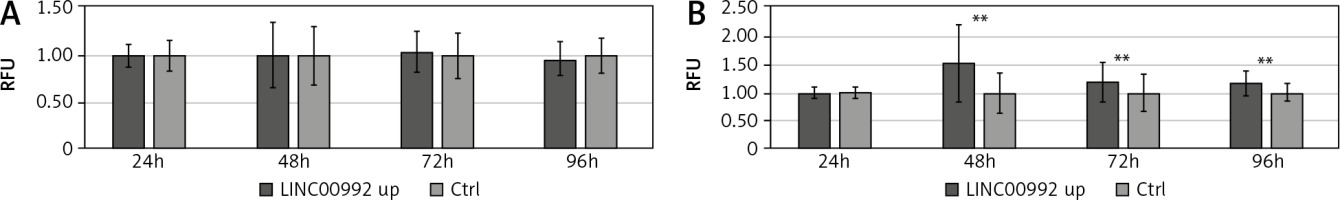
Fig. 6
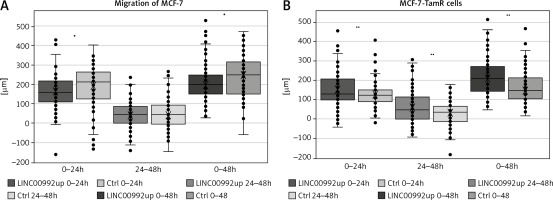
Migration of MCF-7 and MCF-7-TamR cells overexpressing LINC00992 in scratch assays
Representation of cells overexpressing LINC00992 (LINC00992 up) and a control group (Ctrl) receiving an empty plasmid. The experiment was repeated 3 times with 8 replicas each. Within the 24-h and also 48-h interval, LINC00992 overexpressing cells migrated slower than control cells (p < 0.05). However, migration slowed down after 24 hours. For the 24–48-h interval, hardly any differences could be found between the 2 groups.
Proliferation and migration
We further investigated the influence of LINC00992 on cellular activity by determining the proliferation/vitality using the resazurin assay. For this, proliferation and viability were monitored for 4 days in LINC00992-overexpressing MCF-7 and MCF-7-TAM-R cells. Here, MCF-7 cells showed no changes after LINC00992 up-regulation compared with the control group. However, the LINC00992 overexpressing MCF-7-TAM-R cells presented highly significant (p < 0.01) increased fluorescence levels of resorufin compared to the control group at all time points, inferring higher proliferation/viability (Fig. 5).
The scratch assay represents a convenient tool for assessing the cell´s migratory potential. Cells are grown to confluence and then serum-starved to prevent further proliferation. A scratch is applied and determined how fast the cells are able to invade this wound to close it. Using this assay, MCF-7 and MCF-7-TAM-R cells were compared after up-regulation of LINC00992 by the plasmid vector with a respective control group that received the plasmid without insert. Cell migration was monitored for 48 hours, with the travelled distance measured at 24 and 48 hours. The LINC00992 overexpressing MCF-7 cells migrated a distance approximately 30 µm less than the control group after 24 hours (p < 0.05). Over the entire period, cells of the control group migrated approximately 40 µm further than MCF-7 cells after LINC00992 up-regulation (p < 0.01). The interval of 24–48 hours showed less migration and no significant difference. The LINC00992 overexpressing MCF-7-TAM-R cells migrated a total of about 55 µm further than the control group: about 25 µm in the first 24 hours and about 30 µm in the second interval. These differences were highly significant in all time intervals (p < 0.01) (Fig. 6).
Colony-forming assay
The ability of isolated cells to form colonies was examined using colony-forming assays. For this experiment, colonies of LINC00992 over-expressing MCF-7 and MCF-7-TAM-R cells were compared to a counterpart control group. After LINC00992 up-regulation, MCF-7 cells formed about 40% fewer colonies than their control group (Fig. 7). This difference just missed the significance level in the 2-sided t-test (p = 0.055). When comparing the average areas formed by the colonies, no differences were found. The LINC00992 transfected MCF-7-TAM-R cells also formed about 50% fewer colonies than the control group. This difference was nearly significant in the 2-tailed t-test (p = 0.059). Regarding the average area formed by one colony, there were no significant differences between the experimental and control MCF-7-TAM-R groups (Table 3).
Discussion
The molecular mechanisms of acquired tamoxifen resistance are diverse and not conclusively understood, including the contribution of long non-coding RNAs [24]. Cellular models for acquired tamoxifen resistance have been analysed frequently to define tamoxifen-resistance mechanisms. Several deregulated genes identified by these means have been shown to have an impact on the prognosis of breast cancer patients. We have reported on gene expression changes in such a cellular model, focusing on lncRNAs [10], and extended this investigation on microRNAs and eukaryotic initiation factors [39]. We report herein on further studies on LINC00992, a long non-coding RNA that is down-regulated is response to long-term exposure of MCF-7 cells to low doses of 4OH-tamoxifen (10 nM) [10]. This investigation comprises database analysis and, more importantly, manipulation of LINC00992 expression in MCF-7 and the tamoxifen-adapted derivative MCF-7-TAM-R by siRNA and plasmid-based overexpression.
LINC00992 expression among the molecular subtypes of breast cancer and cell lines
Analysis of published gene expression data revealed that LINC00992 is highly expressed in cell lines and breast cancer cells belonging to the oestrogen receptor-positive molecular subtypes luminal A and B [26]. Our database analysis of carcinoma cases revealed the highest expression of LINC00992 in the luminal B subtype. This subtype is clinically more aggressive than the luminal A subtype [40]. However, a high expression of LINC00992 is associated with poorer prognosis and lower median survival only in luminal A carcinomas (Fig. 3). This suggests that high LINC00992 expression is associated with the more aggressive potential of luminal B carcinomas.
Interestingly, the highest LINC00992 expression is found in the luminal A cell line T47D. This cell line is often applied for progesterone receptor (PR) studies [41], and fulvestrant-resistant cell lines have also been generated [42, 43]. One can therefore speculate that high PR expression might be associated with LINC00992 abundance, and it seems promising to investigate this lncRNA in resistant T47D cell lines. A simple explanation for down-regulation by tamoxifen would be a direct regulation via the oestrogen or progesterone receptors. However, ER and PR binding sites seem not to be present in the promoter of LINC00992. Therefore, an indirect regulation via other signals must be postulated [44].
Altogether, LINC00992 is increased in breast cancer and is associated with poor prognosis, as also seen by Li et al. [26]. Thus, high expression of LINC00992 qualifies as a potential prognostic biomarker for breast cancer, particularly for the luminal A subtype. However, further prospective studies should be conducted to evaluate its suitability in greater detail.
Manipulation of LINC00992 expression
We were able to overexpress LINC00992 transiently about 100-fold; however, siRNA transfection resulted in a modest but nevertheless significant down-regulation of 37%. As the down-regulation effect was rather limited and MCF-7-Tam-R showed already decreased expression, we decided to apply this technique only for MCF-7 cells. Concerning the MCF-7-TAM-R cells, we also expected that up-regulation of this down-regulated lncRNA would be more meaningful. We then analysed parameters that can be linked to cancer aggressiveness such as proliferation/viability, migration, and colony forming. Comparing the migration behaviour of the control groups of MCF-7 and MCF-7-TAM-R, it is noticeable that the MCF-7 cells migrated approximately 1.5-fold further than the MCF-7-TAM-R cells. Also, in the proliferation and viability assay, the MCF-7 control group achieved higher fluorescence levels of resorufin than those of MCF-7-TAM-R cells, inferring higher metabolic performance. In the colony-forming assay, the control group of MCF-7 formed about 6 times more colonies than the control group of MCF-7-TAM-R cells. It is therefore noticeable that the MCF-7 cell line appears more vital than the MCF-7-TAM-R cell line. These differences correlated with the morphological appearance of the cells. The MCF-7 cells clustered in cohesive associations with membranous adhesions and mutual shaping of the cell membrane. In addition, the moderately pleomorphic cells formed ductal or tubular duct-like structures [45] (Suppl. Fig. 6). The markedly pleomorphic MCF-7-TAM-R cells grew dissolutely without formation of cell assemblies or organoid structures. Cytologically, the cells presented as polygonal to spindle-shaped with distinctly anisomorphic nuclei (Suppl. Fig. 7).
The suspected metabolic alterations have already been seen in independent tamoxifen-adapted cells [46]. Indeed, these authors found a significant decrease in oxygen consumption and an increase in reactive oxygen species accompanied by a decrease in anti-oxidant defence. Consistent with these data, our group also found a decrease in free glutathione levels in the MCF-7-TAM-R cells [47].
Nevertheless, overexpression of LINC00992 caused a reduced scratch closure in MCF-7 but increased it in MCF-7-TAM-R. Viability of MCF-7 was not affected, whereas it was increased in the tamoxifen-adapted cell line. In both cell lines, the formation of colonies from single cells was reduced. These differences might reflect the different biology of the cells. Interestingly, an up-regulation of LINC00992 seems to have a positive effect on the tamoxifen-adapted cells, so that the cells change towards the higher vitality of MCF-7. Increased migration and proliferation in vitro suggest similar tumour behaviour in vivo. Tumours with high LINC00992 should present with a more aggressive clinic and poorer progression. Our database analysis indeed showed that this was the case, but only for luminal A tumours. Therefore, whether LINC00992 positively or negatively affects viability and migration behaviour in vivo is not conclusively clear and should be analysed in further studies.
Most of the molecular functions of long non-coding RNAs are associated with gene regulation. Because we found that LINC00992 effects migration and proliferation, we further focussed on gene expression changes caused by the manipulation of LINC00992 expression in MCF-7 and MCF-7-TAM-R cells. We applied 2 different methods with different numbers of replicates and genes to determine the effects of manipulating LINC00992 expression. The nCounter® method can be regarded as a screening method (n = 3) including a higher quantity of genes (92), whereas the subsequent qRT-PCR was intended to verify selected results with a higher number of replicates (7–8) and an independent technique. Nevertheless, both methods are accepted for quantification of gene expression. Besides the technical principles, these assays may also detect different parts of the mRNA depending on the oligonucleotides used. Therefore, quantitative results and statistical significances of the results are expected to vary. However, the direction of gene expression changes should be comparable in both methods to attach significance to the results. The 42 gene set contains tamoxifen-regulated and housekeeping genes as well as LINC00992 itself [10]. Here, expression changes should become apparent when LINC00992 has an impact on the establishment of the tamoxifen-related gene expression pattern. Furthermore, a measurement was performed using the Prosigna/PAM50 test set, which is clinically used to determine molecular subtypes and to estimate recurrence risk. This should answer the question of whether LINC00992 can cause changes in the molecular subtype.
Most changes were observed for tamoxifen-related genes. The effected genes were UBD, ZNF462, SIM1, and CEACAM6. In the pam50 panel only one gene (MMP-11) showed a modest but significant change.
The most prominent changes were observed for UBD. This gene is up-regulated in response to 4OH-tamoxifen. It is also known as human leukocyte antigen (HLA)-F adjacent transcript 10 (FAT10), a ubiquitin-like modifier [48]. It has the ability to label its substrates independently of ubiquitin, thus targeting these proteins for degradation by the proteasome [49]. Ubiquitin D is suspected to act as a protooncogene [50]. Its expression is increased in various tumour entities [48] including breast cancer. How the expression of UBD is regulated and its function in malignant tumours remains unclear [50]. High expression of UBD in triple-negative breast carcinomas is associated with poorer prognosis, increased metastasis, and resistance against epirubicin [51]. For instance, the triple- negative carcinoma cell line MDA-MB-231 showed resistance against epirubicin [51]. Likewise, high expression of UBD in MDA-MB-231 correlates with increased sensitivity to cisplatin [52]. This triple-negative cell line displayed a low expression of LINC00992, below the limit of detection in the database analysis (Fig. 1). A correlation between high expression of LINC00992 and cisplatin resistance is suspected [53]. These results suggest that there is a relationship between the expression of UBD and LINC00992 leading to a response to or development of resistance towards cisplatin. Gene expression analysis by qRT-PCR showed significantly decreased relative expression of UBD in MCF-7 and MCF-7-TAM-R when LINC00992 was up-regulated. This result was also evident when MCF-7 cells were measured in the NanoString nCounter®. However, the expression change was only statistically significant in qRT-PCR. Silencing of LINC00992 in MCF-7 cells increased the relative expression of UBD, respectively. However, this effect was only evident when measured in nCounter®, with no statistical significance. The significance of LINC00992 as a possible mediator of tamoxifen, cisplatin, and epirubicin resistance by decreasing the expression of UBD in breast cancer should be further investigated. The mechanism of a possible interaction between LINC00992 and UBD is currently unclear and should be further explored.
During the development of tamoxifen resistance, ZNF462 expression was increased in MCF-7 cells [10]. Up-regulation of LINC00992 in MCF-7 cells decreased the expression of ZNF462. This would be consistent with a regulation via LINC00992. Currently, there is little knowledge on ZNF462 in breast cancer. Nevertheless, GEPIA2 shows that this gene is down-regulated in luminal and HER2-enriched tumours compared to normal tissue. In survival analysis, high mRNA abundance is associated with unfavourable outcome (HR = 1.8, p = 0.012, expression dichotomized at the third quartile) in luminal tumours. Further investigation should therefore be done to verify whether ZNF462 is regulated by LINC00992 and to clarify the possible regulatory mechanism.
Single-minded homologue 1 has been denoted as an “up/down” gene because we found it up-regulated in the early time points and down-regulated at late time points of 4OH-tamoxifen adaption [10], whereas LINC00992 was constantly down-regulated already after one week of incubation. In breast cancer, the SIM1 gene is increasingly methylated, and these tumours have a higher risk of metastasis [54]. This association has also been observed in adenocarcinomas of the lung [55]. In the work of Kim et al. [56], it was shown that hypermethylation of SIM1 in cervical carcinomas resulted in decreased expression of the gene. In regular cervical epithelium, hypermethylation of the gene is not present. By interacting with the DNA methylases DNMT1, DNMT3A, and DNMT3B, LINC00992 may induce methylation of SIM1 and thereby influences its expression. Perhaps LINC00992 induces hypermethylation of SIM1 through the same interaction. The LINC00992 overexpressing MCF-7 cells expressed less SIM1 than controls (log2Fc = –0.68, p < 0.05) (Table 2). siRNA-treated MCF-7 also had modestly down-regulated SIM1 although this difference was not statistically significant. In MCF-7-TAM-R SIM1 expression was reduced after LINC00992 up-regulation, although again this effect was statistically insignificant. The relationship between high expression of LINC00992 and decreased expression of SIM1 should be further investigated, particularly whether the decreased expression of SIM1 results from hypermethylation, which may be affected by LINC00992.
Within the PAM50 gene set only one gene, MMP11, was slightly altered by LINC00992 expression manipulation. Because the PAM50 gene set is clinically applied to determine breast cancer subtypes and thereby estimate the recurrence rate, we conclude that LINC00992 does not influence the subtype of MCF-7 significantly. MMP11 is increasingly expressed by breast cancer, especially in triple-negative subtypes, as well as their metastases, other tumour entities, and even peritumoral connective tissues, such as fibroblast-like cells and adipocytes [57–61]. This increased expression is associated with poor prognosis and tumour progression [62-64].
In summary, LINC00992 manipulation mostly effects the tamoxifen-regulated genes UBD, ZNF462, and SIM1.
The decreased gene expression of UBD and ZNF462 after up-regulation of LINC00992 in MCF-7 matches the increased expression of both genes during the development of a tamoxifen resistance with reduced expression of LINC00992.
Expression of SIM1 was paradoxically shown to be decreased not only during the development of a tamoxifen resistance, with decreased expression of LINC00992, but also during the overexpression of LINC00992 in MCF-7 cells. This might be an artefact caused by massive overexpression. Alternatively, the effect of LINC00992 expression manipulation on SIM1 expression might change after longer treatment as it does under 4OH-tamoxifen exposure. Nevertheless, regulation of SIM1 should be investigated further.
Conclusions
We herein present evidence showing that the ta- moxifen-regulated long non-coding RNA LINC00992 might become a useful unfavourable prognostic biomarker for the luminal A breast cancer subtype. In vitro experiments demonstrated that expression manipulation of this tamoxifen down-regulated RNA caused alterations in viability/proliferation and migratory potential. Furthermore up- or down-regulation of LINC00992 had a significant impact on some tamoxifen-regulated genes, especially UBD/FAT10, ZNF462, and SIM1. The exact regulatory mechanisms of these gene expressions changes remain uncertain and should be elucidated in further investigations, as well as the suitability of LINC00992 as a prognostic biomarker for breast cancer and other tumour entities.