Introduction
In addition to the well-known membrane-bound subcellular organelles in eukaryotic cells, there is now great attention focused on membrane-less cytoplasmic and nuclear structures comprising biomolecular condensates (also called supramolecular assemblies) of proteins and nucleic acids which form functional organelles [1–5]. These condensate structures develop through phase-separation mechanisms in both the cytoplasm and the nucleus [1–5]. Such condensates organize subcellular biochemistry in novel ways and provide spatial segregation of biochemical and signaling pathways. The liquid-like internal nature of these structures assists in increased efficiency of cellular biochemical processes because these allow for co-presence of functionally related proteins and nucleic acids through the process of coordinated phase transitions and their rapid mixing [1–5]. Examples of such membrane-less organelles include the nucleolus, nuclear speckles, P bodies, stress granules, and more recent discoveries such as cytoplasmic condensates of synapsin or of the DNA sensor cyclic GMP-AMP synthase (cGAS) [1–9], and even of viral replication complexes [10, and citations therein]. Overall, these condensates are metastable, changing to a gel or to filaments commensurate with the cytoplasmic environment (temperature, physical deformation, or cytoplasmic “crowding”) [1–10].
Especially relevant to the present discussion is the discovery that transcription factors and RNA polymerase II exist in the nucleus in condensates which efficiently associate with transcriptionally active DNA, often at transcription pause sites [9, 11]. RNA polymerase II subunits and transcription regulatory proteins such as BRD4 and MED1 associate at these punctate condensates [9, 11]. The liquid-like nature of such condensates is typically demonstrated by rapid fluorescence recovery after photo-bleaching (FRAP) experiments as well as by their rapid disassembly (in 15–60 seconds) by 1,6-hexanediol [3, 11, 12]. This rapid sensitivity to hexanediol (3–5%) is now used as a simple test for the liquid-like state of the interior of biomolecular condensates [11, 12].
We recently reported that the unexpected discovery that the human interferon (IFN)-inducible “myxovirus resistance protein A” (MxA), which displays antiviral activity against several RNA and DNA viruses, existed in the cytoplasm largely in membrane-less metastable condensates exhibiting rapid reversible tonicity-driven phase transitions on the time-scale of 1–3 min [13–15]. Exposing Huh7 hepatoma cells expressing GFP-MxA to hypotonic medium rapidly disassembled the cytoplasmic MxA structures within 1–3 minutes. Re-exposure to isotonic medium rapidly reassembled such structures within 1–2 min. Moreover, the GFP-MxA condensates included the DNA sensor cGAS [6]. Functionally, GFP-MxA expression inhibited DNA/cGAS-responsive ISG54-luciferase activity but enhanced the relative fold-inducibility of ISG54-luc by IFN-α, suggesting physical separation between condensate- and cytosol-based signaling pathways. Since regulated phase transitions of biomolecular condensates in the cytoplasm and nucleus are increasingly viewed as critical in diverse cellular functions [1–12], the unique discovery that cytoplasmic MxA condensates underwent rapid and reversible bulk disassembly and then reassembly, each largely completed in a matter of less than 1–3 min, in cells cycled through exposure to hypotonic and then isotonic buffers, was exceptionally striking. Mechanisms driving the observed MxA tonicity-driven condensate phase transitions would include physical events such as a decrease in cytoplasmic crowding [8, 12].
Cytokines and STAT3 in cancer cell biology
It is now well established that a large number of cytokines that signal to the cell interior by activating the transcription factor STAT3 are directly involved in the processes of carcinogenesis and of metastasis [16, 17]. This is especially true in the context of the local tumor microenvironment, where interleukin-6 (IL-6) and activated STAT3 play key roles in cancer progression [16, 17]. Extensive studies of the IL-6/STAT3 pathway in hepatoma cell lines (Hep3B, HepG2), and other cell types, using GFP-STAT3, have revealed the presence of IL-6-induced cytoplasmic and nuclear “bodies” [18–20].
In 2003, Herrmann et al. called attention to GFP-STAT3 nuclear bodies in IL-6-treated Hep3B cells [18]. The appearance of nuclear GFP-STAT3 bodies required IL-6 induction; these contained Tyr-P-STAT3 (PY-STAT3), and, by FRAP, were observed to contain a pool of GFP-STAT3 that was readily mobile [18]. The IL-6-induced nuclear STAT3 bodies also contained the CREB-binding protein (CBP) and histone H4, which are markers for transcriptionally active chromatin. In contrast, these STAT3 nuclear bodies were distinct from the promyelocytic leukemia oncoprotein (PML) bodies. Parenthetically, PML bodies also are now known to be phase-separated condensates [1–5].
In 2007, we called attention to GFP-STAT3 cytoplasmic bodies (we called these “STAT3 sequestering endosomes”) in IL-6-treated Hep3B cells [19]. These cytoplasmic STAT3 bodies required IL-6 induction, contained PY-STAT3, required the Y705 tyrosine in STAT3, and were transient in that these disappeared in 2–3 hours. Moreover, their appearance was blocked by phosphorylation inhibitors such as genistein, staurosporine and indirubin E804, and by the microtubule inhibitor nocodazole [19]. These IL-6-inducible cytoplasmic STAT3 bodies were observed irrespective of the fluorescent tag used (GFP, YFP or DsRed) [19, 20]. Curiously, we were unable to associate any of the known endosome markers with these IL-6-induced cytoplasmic GFP-STAT3 sequestering structures by co-localization assays using immunofluorescence techniques (data not shown). Moreover, extensive attempts to obtain subcellular fractions enriched in such STAT3 sequestering “endosomes” were unsuccessful (data not shown). Critically, this is now no longer surprising in that we note that all such cell fractionation experiments in our hands began with hypotonic swelling of IL-6-induced Hep3B cells followed by Dounce mechanical breakage of the cells (see Fig. 5 below).
Interleukin-6-induced cytoplasmic and nuclear GFP-STAT3 bodies in Huh7 hepatoma cells
We have extended these previous GFP-STAT3 observations of nuclear bodies from the Heinrich laboratory [18] and of cytoplasmic bodies from our laboratory [19, 20], to Huh7 hepatoma cells. Figure 1 illustrates examples of untreated GFP-MxA expressing Huh7 cells as well as those exposed to IL-6 for 15–20 min. The latter cells showed extensive development of cytoplasmic and nuclear GFP-STAT3 bodies.
Fig. 1
Interleukin (IL)-6-induced cytoplasmic and nuclear GFP-STAT3 bodies in Huh7 hepatoma cells. Just subconfluent cultures of Huh7 cells in 35 mm plates that had been transiently transfected with the pGFP-STAT3 expression vector one day earlier were imaged using live-cell microscopy without IL-6 exposure or after exposure to IL-6 (20 ng/ml) for 15–20 min using methods outlined by Xu et al. [19]. Nuc – nucleus, Cytopl – cytoplasm. Scale bar = 25 μm
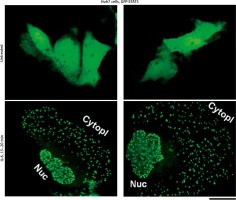
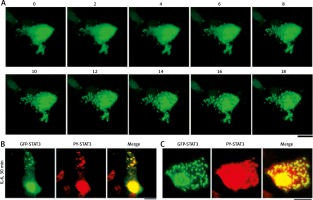
Figure 2A summarizes the rapidly inducible nature of the appearance of cytoplasmic GFP-STAT3 bodies of IL-6-treated Hep3B cells. The cytoplasmic bodies appeared by 10–15 min [19]. Moreover, Figure 2B summarizes the presence of PY-STAT3 in such cytoplasmic (and nuclear) GFP-STAT3 bodies [19]. We note that these GFP-STAT3 structures, cytoplasmic and nuclear, were seen only in cells stimulated with cytokine.
Fig. 2
Association of GFP-STAT3/PY-STAT3 with cytoplasmic structures in interleukin-6 (IL-6)-treated Hep3B hepatoma cells (this figure is an abbreviated version of Figure 1 of Xu et al. [19]). A) Hep3B cells cultured in 6-well plates were transfected with the pGFP-STAT3 construct and imaged 20 hours later using live-cell confocal microscopy. IL-6 (25 ng/ml final concentration) was added immediately after the “0 minutes” frame and the cells were imaged at 15 seconds intervals for the next 18 min. Selected frames from this time-lapse sequence at indicated times in minutes are illustrated. B and C) Hep3B cultures co-transfected with pGFP-STAT3 construct one day earlier were treated with IL-6 for 30 min, fixed with paraformaldehyde and immunostained with anti-PY-STAT3 pAb. The two panels illustrate data from two independent experiments. All scale bars = 25 μm
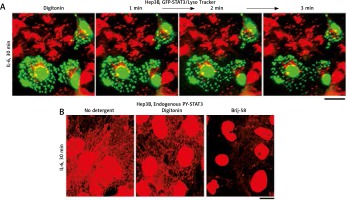
Similar to observations with condensates of cGAS [6] and of MxA [15], the data in Figure 3A (from 2007) show that cytoplasmic GFP-STAT3 bodies were resistant to digitonin [19]. Moreover, Figure 3B shows that even native endogenous STAT3/PY-STAT3 formed punctate structures in the cytoplasm of IL-6-treated Hep3B cells that resisted digitonin, but were disassembled by Brij-58 [19].
Fig. 3
Interleukin-6 (IL-6)-induced GFP-STAT3 and endogenous PY-STAT3-containing cytoplasmic bodies in Hep3B hepatoma cells were resistant to digitonin (this figure is an abbreviated version of Figure 2 of Xu et al. [19]). A) Hep3B cultures transfected with the pGFP-STAT3 construct were first treated with IL-6 for 30 min in the presence of LysoTracker (in red) with GFP-STAT3 in green. These were then sequentially imaged upon treatment with digitonin (50 μg/ml) as indicated. B) Replicate Hep3B cultures were exposed to IL-6 for 30 min and then sequentially to digitonin (50 μg/ml) in ice-cold 0.25 M sucrose/phosphate-buffered saline (sucrose buffer) or Brij58 (0.5% v/v in sucrose buffer), fixed with paraformaldehyde and immunostained for PY-STAT3. All scale bars = 25 μm
Interleukin-6-induced cytoplasmic and nuclear bodies were tonicity-regulated biomolecular condensates
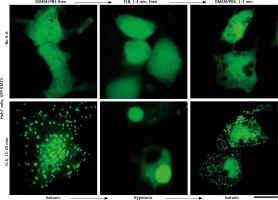
We applied our recent insights into the structure of GFP-MxA condensates in Huh7 cells [14, 15] to the IL-6-induced GFP-STAT3 cytoplasmic and nuclear bodies. Figure 4 shows three independent experiments in which IL-6-induced GFP-STAT3 cytoplasmic and nuclear bodies were disassembled in less than 1 min by exposure to hexanediol. These data provide evidence that the cytoplasmic and nuclear GFP-STAT3 bodies comprised phase-separated biomolecular condensates with liquid-like properties. Even more striking was the discovery summarized in Figure 5 that the integrity of both the cytoplasmic and nuclear bodies was regulated by the tonicity of the culture medium. A switch to hypotonic ELB medium led to disassembly of both cytoplasmic and nuclear GFP-STAT3 bodies within 1–3 min. Re-exposure to isotonic medium led to reassembly of cytoplasmic and nuclear GFP-STAT into discrete structures – but different from the starting structures. These observations recapitulate the tonicity-driven disassembly and reassembly of GFP-MxA in Huh7 cells observed by us [14, 15] and suggest that cytoplasmic “crowding” [4, 8] may be a likely mechanism regulating these phase separations.
Fig. 4
Interleukin-6 (IL-6)-induced cytoplasmic and nuclear GFP-STAT3 bodies are phase-separated condensates. Huh7 cultures in 35 mm plates transfected with pGFP-STAT3 vector one day earlier were treated with IL-6 (20 ng/ml) for 15–20 min. Live cells showing IL-6-induced cytoplasmic and nuclear bodies 15–20 min later identified, then exposed to hexanediol (5%) in phosphate-buffered saline (PBS) and imaged immediately thereafter. Figure illustrates three independent experiments. Scale bar = 25 μm
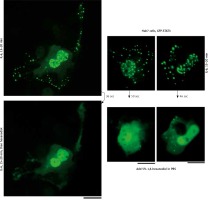
Fig. 5
Culture-medium tonicity drives disassembly and reassembly of interleukin-6 (IL-6)-induced GFP-STAT3 cytoplasmic and nuclear condensates in Huh7 cells. Huh7 cultures in 35 mm plates transfected with pGFP-STAT3 vector one day earlier were left untreated with cytokine (upper row) or treated with IL-6 (20 ng/ml) for 15–20 min (lower row) and imaged in isotonic medium (DMEM/PBS; approximately 325 milliosmolar), then switched to hypotonic medium (erythrocyte lysis buffer (ELB), approximately 40–50 milliosmolar) for 3 min, and then back to isotonic medium (DMEM/PBS) as indicated. Scale bar = 25 μm
Practical implications
The discovery that the standard hypotonic buffer used by many investigators for cell swelling (as is typically the first step prior to mechanical cell breakage in cell-fractionation protocols) led to rapid and marked disassembly of IL-6-induced cytoplasmic and nuclear GFP-STAT3 condensates (Fig. 5) has implications for interpreting hundreds of studies of the biochemistry of STAT3. We show by the data in Figure 5 that hypotonic cell swelling introduces a hitherto unrecognized limitation into all such studies – there occurs rapid disassembly of cytoplasmic protein condensates under such fractionation conditions.
The recognition that IL-6-induced GFP-STAT3 existed, at least in part, in the cytoplasm and nucleus in biomolecular condensates highlights the possibility of cross-talk between STAT3 and other signaling pathways. Herrmann et al., already in 2003 [18], pointed to the co-association of PY-STAT3 with CBP and histone H4 in IL-6-induced nuclear bodies (which we now identify as “biomolecular condensates”). In line with our previous data on MxA condensates which included co-condensation with cyclic GMP-AMP synthase (cGAS) [15], we suggest that cytoplasmic condensates, such as of GFP-STAT3, might also allow for the physical/spatial segregation of signaling pathways in the cytoplasm as well as cross-talk with other novel signaling mechanisms.
“STAT-masking”: another possible example of IL-6-induced wt p53-dependent STAT3 phase transition in hepatoma cells
In 1997–1998 we reported the curious phenomenon wherein bulk cytoplasmic STAT3 and STAT5 in Hep3B cells expressing wild-type p53 and then treated with IL-6 transiently lost their ability to be detected by immunofluorescence methods, even though there was no loss of the respective proteins [21, 22]. This phenomenon, dubbed “STAT-masking,” was evident in less than 30 min after IL-6 exposure and lasted 2–3 hours [21, 22]. IL-6-induced STAT-masking was selective in that it was observed for STAT3 and STAT5, but not STAT1. STAT-masking required IL-6-induced Tyr phosphorylation as well as wild-type p53 expression and protein synthesis for at least 6–8 hours prior to its successful elicitation [21, 22]. This loss of immunoaccessibility of STAT3 and STAT5 may represent a novel bulk phase transition mechanism dependent upon a p53-induced cellular protein. That exposure of Hep3B cells to IL-6 leads to the development of large (1-2 MDa) complexes of GFP-STAT3 has been inferred from fluorescence correlation spectroscopy of the cytoplasm of live cells [23].
Conclusions
We provide a novel reinterpretation of previous incompletely understood cell biology data in the STAT3 signaling field with respect to IL-6-induced activation of this transcription factor in hepatoma cells and the formation of cytoplasmic and nuclear STAT3 bodies. We now recognize that the IL-6-induced GFP-STAT3/PY-STAT3 cytoplasmic and nuclear bodies represent phase-separated biomolecular condensates which disassembled rapidly in the presence of hexanediol. This IL-6-induced generation of these cytoplasmic and nuclear STAT3 bodies represents an example of a cytokine-triggered phase transition which is Tyr phosphorylation dependent and which occurred in less than 10–15 min. Moreover, we observed that these STAT3 condensates showed rapid tonicity-driven phase transitions. The new insights extend the concepts of phase-separated biomolecular condensates to the cytokine-induced activation of STAT transcription factors in cancer cell biology, and point to a novel underlying biochemistry.








