Purpose
Accurate delineation of both target organs and organs at risk (OARs) is crucial for the planning of modern radiotherapy [1]. As a form of precision treatment, low-dose-rate (LDR) brachytherapy delivers high radiation doses to the anatomy of each target volume, while sparing surrounding normal tissues. Interstitial brachytherapy after parotid cancer surgical resection, can be used to deliver high-conformity radiation doses to target volumes, and it provides a high local control rate with few side effects [2]. OARs segmentation is currently manually performed, and involves screening of hundreds of computed tomo-graphy (CT) imaging slices. As such, this process is not only tedious and time-consuming, but is also prone to subjectivity and inter-operator variability.
Automated segmentation has been developed to address these challenges. Various deep learning-based systems methods have been derived from previous statistical models or atlas-based models [3]; however, the presence of post-surgical adhesions, which can compromise soft tissue boundaries on imaging, remains a challenge in adjuvant radiotherapy. Currently available algorithm-derived volumetric segmentation techniques tend to require significant manual editing, and fail to provide significant improvement in clinical workflow [4]. Advancements in deep learning technologies in medical image classification and segmentation have gained substantial attraction and success in recent years, particularly convolutional neural networks (CNN) [5]. Multiple CNN-based models have been proposed in the past decade, such as fully convolution networks (FCNs) [6], DeepLab system [7], and U-net [8].
With the adoption of symmetrical expansion paths and skip connections, U-net fuses low- and high-level feature maps, and effectively overcomes imaging noise and blurred boundaries. U-net and its’ variants have thus been widely used for segmentation during medical imaging, especially in the field of radiotherapy. Recently, the nnU-Net method, which builds on the architecture of U-net, has been considered the best baseline deep learning-based model for medical segmentation, owing to its’ automaticity in all stages, from pre- to post-processing of each given image dataset, removing the need for manual tuning [9]. Due to the scarcity of studies on the use of automated segmentation models in parotid cancer, we thereby proposed a two-step 3D nnU-Net-based automated OARs segmentation model for brachytherapy in parotid gland cancer, and evaluated its accuracy by comparison with gold standard expert manual segmentation.
Material and methods
Clinical imaging data
A total of 200 patients with parotid gland carcinoma, who underwent surgical resection followed by adjuvant iodine-125 (125I) seed with surface radioactivity of 18.5 MBq and air-kerma strength of 0.635 μGy• m2• h−1 per seed (model 6711; Beijing Atom and High Technique Industries Inc., Beijing, China; t1/2, 59.6 days; energy level, 27.4-31.4 keV; tissue penetration capacity, 1.7 cm) with activity of 22.2 to 29.6 MBq (range, 0.6-0.8 mCi) implantation brachytherapy [10] at the Peking University Stomatology Hospital between 2017 and 2021 were included in the study. For each patient, treatment planning was performed using a brachytherapy planning system (BTPS; Beijing Astro Technology Ltd. Co., Beijing, China), based on CT images taken within 4 weeks post-surgery.
Images were acquired in 512 × 512 matrix size and 2 mm slice thickness. All images were cut in the sagittal position, and only data on the side of cancer were retained for the study. Six OARs were defined, including the auricula, condyle process, skin, mastoid process, external auditory canal, and mandibular ramus. It should be noted that some OARs in parotid brachytherapy have a large range, such as the skin and mandible. Nevertheless, collateral damage to normal tissues in brachytherapy is likely to be confined to the vicinity of clinical target volume (CTV), it was not possible to outline the whole organ. Regions of interest (ROIs) were limited to 17 mm from the residual parotid tissues, in accordance with inherent irradiation range of 125I seeds. Collected datasets were randomly divided into a training set of 70% of cases (n = 140) to establish and train the proposed model, a validation set of 15% cases (n = 30) for held-out validation, and a test set of 15% of cases (n = 30) to evaluate the performance of the final prediction models.
Two-stage diagnostic labeling was performed by two experienced radiation oncologists (reader 1 and reader 4, with 5 years’ and 10 years’ experience in parotid cancer treatment planning, respectively) in consensus. Prior to this, training was performed on five cases unrelated to the study to allow for discussion of segmentation procedures and development of consensus criteria.
In the first stage, the radiologist with 5 years of experience manually labeled 6 organs using an open-source software package (ITK-SNAP version 3.4.0; http://www.itksnap.org) [11]. All 200 patients’ OARs were re-delineated for the purpose of this research. In the second stage, contour sets were reviewed by a senior radiologist and modified as necessary, to maintain consistency. Manual delineation were used as gold standard for training and testing.
The present study was approved by the Institutional Review Board. All image data were obtained retrospectively, and were anonymized and de-identified prior to analysis.
Deep learning two-step segmentation model
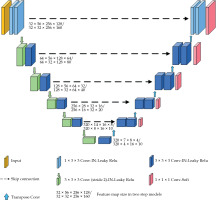
The architecture of 3D nnU-Net model is shown in Figure 1. As an out-of-the-box tool, pre-processing strategies and training hyperparameters were automatically adapted. The model was comprised of an encoder and decoder path, with skip connections in between. Both paths consisted of repeated blocks of two convolutional layers, each followed by an adaptive layer-instance normalization layer and a leaky rectified linear units. In the encoder, down-sampling was done by strode convolution, while in the decoder path, up-sampling was performed via transposed convolution. In the decoder, feature maps after each convolutional block were created by convolution with a 1 × 1 × 1 kernel, and SoftMax function was applied to the output for deep supervision. Initial feature maps were set to 32, and were doubled during down-sampling to a maximum of 320.
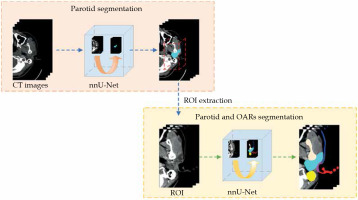
A two-step approach was designed to allow for coarse-to-fine segmentation, as shown in Figure 2. The first model was trained to segment the parotid gland only, while the second was trained to segment the pre-defined ROIs. As mentioned, the input of the second model was created by expanding 17 mm from the position of the segmented parotid gland identified in the first model. The test dataset was used as the input of each model.
A total of 170 patients’ CT scans were used for training and validation. Based on the self-configurable strategy of nnU-Net, all images were automatically pre-processed by re-sampling, normalization, data augmentation (mirroring, rotation, and scaling), and cropping prior to network input. Input volumes of each model were resized to 56 × 256 × 128 and 32 × 256 × 160 pixels, respectively.
Loss and training details
The model was trained with deep supervision [12]. For each output, a corresponding down-sampled ground-truth segmentation mask was used for loss computation. Loss (L) was defined as follows (Equation 1):
where αd and βd are the weight coefficients, and were assigned as 1 and 1; and where Ld represents the loss of d-th layer in deep supervision, d ∈ (1,2,3,4,5).
Loss function (Ld) was defined as the sum of dice loss (Ldice) [13] and cross entropy loss (Lce) [14] as follows (Equation 2):
where yni and pni represent the ground truth segmentation and the predicted probability of ith pixel of nth class, respectively. The pixel and class numbers were denoted as I and N, respectively.
Training of the model was performed using stochastic gradient descent (SGD) optimizer with Nesterov momentum. Initial learning rate was set at 1e-3, and batch size was set to 2, with each epoch defined as 250 training iterations. Maximum epoch was set to 1,000. The proposed model was implemented on a single GeForce RTX 3090 (NVIDIA) graphics processing unit of 24 GB. Codes were developed with Python version 3.7.9, while the model was developed using PyTorch version 1.8.0.
Quantitative evaluation metrics
The accuracy of the model was qualitatively evaluated against gold standard manual contours in terms of dice similarity coefficient (DSC), Jaccard index, 95th-percentile Hausdorff distance (95HD), and precision and recall.
DSC describes the degree of volumetric overlap between ground truth (A) and deep learning-based segmentation (B) (Equation 3). Values 0 and 1 corresponded to no and complete overlap, respectively [15].
The Jaccard index is an algorithm performance metric for measuring similarity between datasets. It was defined as the cardinality of intersection divided by the cardinality of union of the A and B sets (Equation 4) [16].
Hausdorff distance was a distance-based metric, and measures dissimilarity between two datasets [17]. It was defined as the largest value of all distances from the point in one set to the closest point in the other. Lower HD values imply higher segmentation accuracy. 95HD was applied in our study because of high susceptibility of HD to noise and local outliers.
The precision and recall were based on true positives (TP), false negatives (FN), and false positives (FP) [18].
The precision was defined as the ratio between true positives and the total number of possible positives (true positives + false negatives). The precision rate was presented in Equation (5).
The recall was defined as the ratio of true positives in the data to true positives and false negatives. The recall rate was represented in Equation (6).
Data were verified with the Kolmogorov-Smirnov test to determine whether they were approximately normally distributed. For normally distributed data, the paired Student’s t-test was used to compare the two-step and segmentation-only models. For non-normally distributed data, the Wilcoxon test was used. Statistical significance was set at two-tailed p < 0.05.
Subjective validation
For qualitative analysis, all tested datasets were blindly reviewed by two senior oncologists with > 20 years of experience. Segmentation quality was graded using a 4-point scoring system, with 0 points meaning severe defect, presence of large and obvious errors; 1 point meaning moderate defect, presence of minor correctable errors; 2 points meaning mild defect, presence of clinically insignificant errors; and 3 points meaning precise, no editing required.
Dosimetric evaluation
To assess dosimetric impact of our proposed model, brachytherapy plans on all test cases were made. Then, dose-volume histogram (DVH) of OARs between two-step model and manual expert segmentation was calculated. Difference of Dmean was determined to access dosimetric effects of each segmentation method. OARs dosimetric metrics were analyzed using Spearman’s rank tests (p = 0.05).
Results
Accuracy of residual parotid tissue segmentation
The accuracy was demonstrated in the first segmentation stage of our proposed model for residual parotid tissue, with DSC of the validation dataset reaching 0.87.
Accuracy of OARs’ segmentation
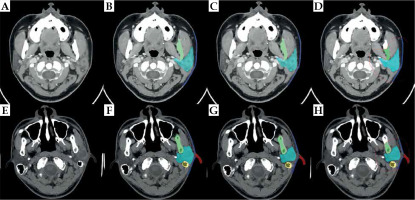
Figure 3 show the best and challenging scenarios obtained with manual expert segmentation (B and F), the segmentation-only model (C and G), and the proposed two-step segmentation model (D and H) in two representative patients. Successful delineation of the six OARs was observed, with good visual agreement across all 3 modalities.
Fig. 3
The best and challenging scenarios in the model. Figures (A-D) show the best scenario. A) A slice obtained from a 3D parotid CT image; B) Result from manual expert segmentation; C) Segmentation-only model; D) The two-step segmentation model. Figures (E, F) show the challenging scenario. E) A slice obtained from another 3D parotid CT image; F) Result from manual expert segmentation; G) Segmentation-only model; H) Two-step segmentation model
Statistical results of the two-step segmentation and segmentation-only models are presented in Table 1. Lower 95 HD and higher DSC values were observed in our proposed model.
Table 1
Statistical evaluation results of four different kinds of autosegmentation models
Mean DSC values of > 0.80 were observed in all OARs, except for the skin and external auditory canal, suggesting the reliability and applicability of our proposed model for the automated segmentation of parotid cancers. Among all structures, the best segmentation results were obtained for the mandibular ramus and condyle processes, with mean DSC values of 93.19% and 91.48%, respectively, and relatively high contrast and clear boundaries were observed in post-operative CT images.
Quantitative data between the two-step and segmentation-only models were compared using the paired Student’s t-test or Wilcoxon test. Statistical significance was set at two-tailed p < 0.05. It can be seen from the data in Table 1 that there were differences in the statistical data between the two-step and segmentation-only models, and most of the data comparisons were statistically significant (p < 0.05).
Oncologist evaluation
The results of the qualitative analysis are demonstrated in Table 2. Automated delineation using the two-step model was deemed clinically acceptable by the two senior oncologists.
Segmentation time
The average time for OARs segmentation with our two-step model was 50.90 s, of which 4.47 s were spent on slice classification. In contrast, the expert manual segmentation required over 20 minutes.
Dosimetric impact
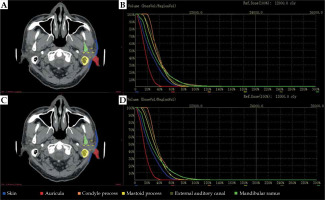
We performed experiments to examine the impact of contours obtained with the proposed automatic segmentation method. Figure 4 shows DVHs for all six OARs of an exemplary patient. We calculated the difference of Dmean on OARs segmented from manual segmentation by expert oncologists and two-step automated segmentation approach for all 30 plans. Group differences were assessed by calculating the dose, with a mean, standard deviation, and corresponding p-value. The mean and standard deviation of dose variables computed over these two groups are presented in Table 3, along with the corresponding p-values. Dosimetric metrics showed p-values larger than 0.05, indicating for all OARs that there were no obvious differences between the dosimetric metrics of these two segmentation methods.
Table 3
The mean and standard deviation (SD) of Dmin differences and p-value calculated with organs at risk segmented by manual expert and two-step segmentation model for all 30 plans
| Mean (Gy) | SD (Gy) | P-value | |
|---|---|---|---|
| Auricula | 5.52 | 12.14 | 0.266 |
| Condyle process | 2.69 | 4.31 | 0.547 |
| Skin | 9.71 | 17.55 | 0.062 |
| Mastoid process | 3.37 | 3.81 | 0.054 |
| External auditory canal | 8.92 | 17.10 | 0.120 |
| Mandibular ramus | 2.53 | 3.34 | 0.619 |
Comparison with other methods
We compared our proposed method with several deep learning-based 3D segmentation methods, including 3D U-Net [19], V-Net [20], and nnU-Net (segmentation-only model). To ensure objectivity, we used the same training framework for the appeal method as our proposed method, including the same data augmentation, pre-processing, post-processing, etc. The data were analyzed in terms of DSC, Jaccard index, 95 HD, and precision and recall.
From Table 1, it can be seen that the two-step approach we proposed was better than the others method in each indicator. Among them, for the skin with a large range, our method was significantly superior to the others, 7% higher than 3D Unet and 2% higher than nnU-Net of the one-step method with mean DSC values.
Discussion
The parotid gland is considered an important organ at risk in head and neck cancer and whole brain radiotherapy [21]; however, studies on OARs in brachytherapy for parotid gland cancer are currently lacking. OARs for parotid cancer patients with external beam radiation therapy have included the contralateral parotid, eyes, lenses, optic nerve, and spinal cord [22]. Owing to the steep gradient in brachytherapy, small size of OARs and similar density of surrounding tissues render segmentation difficult. Moreover, tissue adhesions and subsequent blurring of soft tissue boundaries following surgical resection can further challenge the segmentation process for adjuvant brachytherapy in parotid gland cancer.
The present work proposed a deep learning-based two-step automated organs at risk segmentation technique, and evaluated its performance for brachytherapy planning in parotid gland cancer. Six OARs were identified, including the auricula, condyle process, skin, mastoid process, external auditory canal, and mandibular ramus. Accurate automated segmentation results, with close agreement to those of gold standard manual segmentation, were observed, indicating mean DSC values of 0.88, 0.91, 0.74, 0.89, 0.74, and 0.93, respectively. The entire segmentation process with our model took approximately 50.90 s, while manual segmentation performed by experienced oncologists required over 20 minutes. The potential of our model in improving the efficiency of the segmentation process was thus shown. While DSC is a widely adopted metric in assessing segmentation quality, it is highly sensitive to the size of evaluated item [23]. For adequate statistical evaluation, qualitative validation by experts in this field was considered, who subsequently deemed the two-step automated segmentation approach clinically acceptable. Therefore, our overall results demonstrated the potential of our proposed model as an accurate and efficient segmentation method in clinical practice.
The ranking in descending order of the segmentation results of the six OARs include the mandibular ramus, condyle process, mastoid process, auricula, external auditory canal, and skin, which seemed to be related to the degree of complexity and variation. The mandibular ramus and condyle process have relatively simple shapes, and clean boundaries and strong contrast allow an easy segmentation from image background. By contrast, automatic segmentation is challenging in external auditory canal and skin segmentation due to the complex background texture and large variation in size, shape, and intensity. Occasionally, it is difficult to distinguish the boundaries between residual gland and OARs, depending on initial tumor extent and gross residual diseases. Blurred and low-contrast surgical areas may lead to fuzzy segmentation from automatic segmentation models. In these cases, OARs segmented by the model had relatively poor consistency with those of the radiation oncologists; however, as intra- and inter-observer variability in the quantification of the same structure occur, it was necessary to conduct subjective validation in addition to quantitative evaluation. Numerous examples, in which cases with lower quantitative agreement were still judged as clinically acceptable.
The high variability in architecture and morphology of the parotid gland post-operation often results in heterogeneity in the range of ROIs among different individuals, which can be a major challenge in manual segmentation. As such, automated segmentation methods have received great attraction to enable personalized ROI contouring. We achieved a two-step approach using deep learning, which focused on ROI, and obtained greater accuracy even though the salient region was small when compared with the method that extracted global features of the entire image for estimating the overall quality. As the irradiation only affects a very localized area around the radiation sources in brachytherapy [24], delineation was not necessary for tissues outside the region of interest, which increased noise and artifacts, and potentially contributed to the most uncertainty in radiotherapy planning [25]. Although the region of interest was delimited at 1.7 cm edge of the post-operative residual parotid gland, radiation oncologists who delineate OARs tend to expand the scope to avoid potential omission. If the ROI is labeled accurately on the network method, the accuracy will be reflected in the segmentation. Moreover, as the original CT images contains a large background that carries irrelevant, noisy, and redundant features, this two-step segmentation step method not only improves the segmentation performance by removing the irrelevant (or less relevant) features, but also reduces the computational complexity by decreasing the spatial size of input volume. In this study, ROIs were limited to 17 mm from the residual parotid tissues, in accordance with the penetration depth of selected 125I seed of 17 mm. Higher consistency can be achieved by selecting smaller ROIs.
Due to its influence on tumor control and the risk of radiation-induced toxicity, OARs’ contouring is crucial for radiotherapy planning. Current gold standard manual segmentation is, however, laborious and time-consuming [26]. Increasing attention has been placed on the use of deep learning-assisted models for automated segmentation in brachytherapy [27, 28]; however, to our knowledge, such models have not been utilized in the head and neck region. Precise delineation and prevention of severe radiotherapy-related complications are particularly crucial in head and neck cancers, given the complexity of anatomical structures and clinical emphasis on maintaining aesthetics [29]. At present, the majority of research on automated delineation techniques in the head and neck region has revolved around external radiation therapy. In a study by van Dijk et al., a 2D U-Net network model using CT images of 589 patients was utilized for the segmentation of the parotid gland, which obtained DSC values of 0.81 ±0.08 [30]. Dai et al. proposed a deep learning-based head and neck multi-organ segmentation method using magnetic resonance images of 60 patients, and obtained DSC values of 0.85 ±0.06 and 0.86 ±0.05 in the left and the right parotid glands, respectively [31]. Our results were in line with those of the aforementioned studies. In the current study, surrounding tissues, such as the mandible and skin, were delineated as OARs, but there is a scarcity of similar research data entries for these structures for comparison.
There were several limitations in our study. The six OARs included were selected based on our institutional protocol for parotid brachytherapy, which overlooked other organs, including the masseter and sternocleidomastoid muscle. Therefore, future research with inclusion of these organs is warranted. Due to limited computing power, images with 2 mm slice thickness were acquired. Given that images with a lower slice thickness would provide more and better information, it is possible to acquire better segmentation results using thinner CT slices; however, this will lengthen the segmentation time. We will consider this point in a follow-up research. Thirdly, another potential limitation is that this was a single-center study, which makes the accuracy of the cases out of our patients’ data open to question.
Conclusions
The high variability in architecture and morphology of the parotid gland post-operation often results in heterogeneity in the range of ROIs across different individuals, which can be a major challenge in manual segmentation. With deep learning-based segmentation techniques, the ROI was the first extracted and fed into the system, allowing for a focus on anatomically relevant regions to achieve segmentation of greater accuracy in our proposed model. This approach thereby carries the potential in expediting the treatment planning process of brachytherapy for parotid gland cancers.