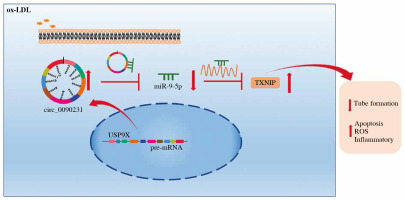
Introduction
As a chronic inflammatory disease, atherosclerosis (AS) has been considered to induce severe clinical complications, such as cerebral infarction, cardiovascular disease, and peripheral vascular diseases [1, 2]. Currently, it is reported that aberrant endothelial cell injury is one of the leading causes of artery atheromatous plaque, which eventually leads to the progression of atherosclerosis [3, 4]. Of note, numerous researchers have discovered that oxidized low-density lipoprotein (ox-LDL) is able to trigger the apoptosis of endothelial cells and is considered as an important risk factor for AS [5, 6]. Hence, exploring the molecular mechanisms by which ox-LDL induces endothelial cell damage is vital for the effective treatment of AS.
Over the past decade, several underlying therapeutic agents for repression of endothelial cell injury have been discovered, containing non-coding RNAs (ncRNAs) [7, 8]. As a novel class of ncRNAs, circular RNAs (circRNAs) have drawn great attention due to their unique feature of a loop covalently closed by combining the 5'-end and 3'-end [9]. More recently, the biological function of circRNAs has received interesting attention in diverse human diseases, including AS [10]. For example, Wang et al. reported that circ_0124644 could aggravate ox-LDL-caused endothelial injury in human umbilical vein endothelial cells (HUVECs) through hindering proliferation and boosting apoptosis [11]. Similarly, Chen et al. found that the silencing of circ-BANP could block cell growth, metastasis, angiogenesis, and promoting apoptosis and inflammation of HUVECs by regulating thioredoxin-interacting protein (TXNIP) via miR-370 [12]. Interestingly, microarray analysis revealed the upregulation of circ_0090231 in ox-LDL induced HUVECs, implying that circ_0090231 might be deemed as an important regulator in AS [13]. However, its precise role and underlying mechanism in AS are still unclear.
At present, circRNAs have been described as competing endogenous RNAs (ceRNAs) that sequester their target microRNAs (microRNAs) away from mRNAs, thereby altering down-stream gene expression [14, 15]. Herein, circ_0090231 was viewed to possess some binding sites with miR-9-5p in HUVECs. Also, relevant literature has reported that miR-9-5p could suppress the formation of vulnerable plaque and vascular remodeling in AS [16, 17]. Thus, the purpose of this study was to determine whether circ_0090231 could regulate endothelial cell injury by interacting with miR-9-5p in AS.
Material and methods
Cell culture and treatment
Under a moist incubator with 5% CO2 at 37°C, human umbilical vein endothelial cells (HUVECs; Procell, Wuhan, China) were grown in Dulbecco’s modified Eagle’s medium (DMEM; PAN Biotech, Aidenbach, Germany) with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA). For verifying the stability of circ_0090231, HUVECs were treated with 3 U/µg of RNase R (Epicenter Technologies, WI, USA) 15 min at 37°C or co-cultured with 2 mg/l of actinomycin D (Invitrogen), respectively.
For ox-LDL treatment, HUVECs were second passaged with confluence of 60-70%, followed by exposure with ox-LDL (Sigma-Aldrich, St. Louis, MO, USA) dissolving dimethyl sulfoxide (DMSO; Sigma-Aldrich).
Real-time quantitative polymerase chain reaction
Based on the supplier’s direction of Trizol reagent (Invitrogen), total RNAs from HUVECs were isolated, followed by synthesizing cDNA using Prime Script RT Master Mix (TaKaRa, Dalian, China). After that, real-time quantitative polymerase chain reaction (RT-qPCR) detection was performed on an ABI 7900 System (Applied Biosystems) with primers, the synthesized cDNA, and a SYBR Green PCR kit (TaKaRa). After normalization with the reference to expression of GAPDH or U6, the relative expression levels were analyzed referring to the 2–ΔΔCt method. Primers are presented in Table 1.
Table 1
Sequences of primers for RT-qPCR used in this study
Cell viability
After treatment of ox-LDL (0, 25, 50, and 75 µg/ml) for different time points, HUVECs were supplemented with 10 µl of 3-(4,5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT, Sigma-Aldrich) for 4 h, followed by the introduction of DMSO (Sigma-Aldrich). Finally, the absorbance value at 490 nm was determined using a microplate reader (ELX808; Biotek, Winooski, VT, USA).
Cell transfection
For circ_0090231 stable expression, HUVECs were infected with sh-circ_0090231, oe-circ_0090231, and their controls (sh-NC or vector), followed by selecting with puromycin. And lentiviral vectors were acquired from Genechem (Shanghai, China). Meanwhile, 30 nM of miR-9-5p mimic, miR-9-5p inhibitor, miR-NC mimic, miR-NC inhibitor, and 200 ng of pcDNA-TXNIP (TXNIP) and pcDNA from RiboBio (Guangzhou, China) were respectively transfected into HUVECs, according to the instruction guidelines of Lipofectamine 3000 (Invitrogen).
Tube formation assay
Angiogenesis ability of HUVECs was assessed by tube formation assay. Generally, after ox-LDL treatment, 5 × 104 HUVECs were introduced into 96-well plates, which were per-coated with Matrigel (BD Biosciences, Heidelberg, Germany). Following incubation for 48 h, a light microscope (Olympus, Tokyo, Japan) was used to analyze the number of branches in this assay.
Cell apoptosis assay
In this assay, the detection of HUVEC apoptosis was carried out using a commercial Annexin (V-fluorescein isothiocyanate) V-FITC/Propidium Iodide (PI) Apoptosis kit (Bender Med System, Vienna, Austria). In short, treated HUVECs were re-suspended in binding buffer containing 5 µl of Annexin V-FITC and 10 µl of PI, followed by analysis using FACScan flow cytometry (BD Bioscience).
Western blot assay
In this assay, HUVEC lysates were prepared with RIPA lysis buffer (Beyotime, Nantong, China), according to the operation manual. After immunoblotting, the proteins were transferred onto nitrocellulose membrane (Millipore, Molsheim, France), which were hybridized with primary antibodies: Bcl-2 (ab194583, 1 : 1000), Bax (ab32503, 1 : 1000), TXNIP (ab188865, 1 : 1000), and GAPDH (ab8227, 1 : 1000) at 4°C all night. After incubation with secondary antibody (ab205178, 1 : 10,000), the immunoreactive bands were detected using an ECL detection kit (Santa Cruz Biotechnology, Santa Cruz, CA, USA). All antibodies were obtained from Abcam (Cambridge, MA, USA).
Measurement of MDA, LDH, and SOD
After 75 µg/ml ox-LDL treatment for 48 h, HUVECs were harvested and lysed. Then the cell extract was utilized for the determination of malondialdehyde (MDA), lactate dehydrogenase (LDH), and superoxide dismutase (SOD) activity, as per the user’s guidebook of the MDA kit (Beyotime), LDH kit (Beyotime), and SOD kit-WST (Dojindo, Kumamoto, Japan).
Enzyme-linked immunosorbent assay (ELISA)
In this assay, after ox-LDL treatment, the culture medium of HUVECs was collected and detected, followed by measurement of tumor necrosis factor α (TNF-α), interleukin 1β (IL-1β), and interleukin 6 (IL-6) using enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems, Minneapolis, MN, USA).
Dual-luciferase reporter assay
Based on the analysis of starBase (http://starbase.sysu.edu.cn) software, the underlying site between miR-9-5p and circ_0090231 or the TXNIP 3' untranslated region (3'UTR) was inferred. The circ_0090231 or TXNIP 3'UTR wild-type (WT) reporter vectors (WT-circ_0090231 or WT-TXNIP 3'UTR) and the site-directed mutant (MUT) vectors (MUT-circ_0090231 or MUT-TXNIP 3'UTR) were provided by Genechem. Then, these vectors were respectively transfected into HUVECs with miR-9-5p mimic or miR-NC mimic, followed by determination using a dual- luciferase reporter assay kit (Promega, Madison, WI, USA).
RNA immunoprecipitation
In this assay, the interaction between circ_0090231 and miR-9-5p was assessed by RNA immunoprecipitation (RIP) assay. In general, HUVECs transfected with miR-9-5p mimic or miR-NC mimic were lysed in the complete RIP lysis buffer (Millipore). After that, cell lysates were incubated with Argonaute 2 antibody (Ago2; Millipore) and a negative control IgG (Millipore) before treating magnetic protein A/G beads for 2 h. After treatment with proteinase K, the RNA was purified for the detection of circ_0090231 using RT-qPCR assay.
Results
Circ_0090231 expression was increased in ox-LDL-treated HUVECs
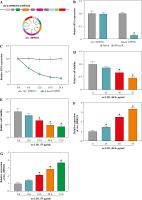
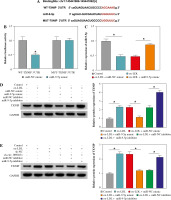
First of all, our analysis suggested that circ_0090231 was generated from 3-12 exons of the ubiquitin-specific protease 9X (USP9X) gene, and the ends of exon 3 and exon 12 were back-spliced to form a circular structure (Fig. 1A). Subsequently, RNase R (a highly processive 3' to 5' exoribonuclease) and actinomycin D (an inhibitor of transcription) were utilized to evaluate the stability of circ_0090231. As presented in Figure 1B, RNase R treatment apparently reduced the expression level of linear USP9X mRNA, whereas it had almost no impact on circ_0090231 expression in comparison with the untreated group. Moreover, the half-life of the circ_0090231 transcript exceeded 24 h, suggesting that this isoform is more stable than the linear USP9X mRNA transcript in HUVECs (Fig. 1C). Then, to prove the effect of circ_0090231 on AS, an in vitro model of AS was established in HUVECs stimulated with ox-LDL. As displayed in Figure 1D, E, cell viability was gradually reduced in HUVECs after treatment of 0-75 µg/ml ox-LDL for 48 h and 75 µg/ml ox-LDL for 0-72 h. Of interest, the upregulation of circ_0090231 under the same condition was observed in HUVECs (Fig. 1F, G), suggesting an underlying role of circ_0090231 in ox-LDL-triggered damage of HUVECs. The 75 µg/ml ox-LDL treatment for 48 h was chosen for further functional analysis.
Fig. 1
Circ_0090231 was elevated in ox-LDL-treated HUVECs. A) Schematic illustration showing uSP9X exons 3-12 circularization to form circ_0090231. B, C) Relative levels of circ_0090231 and uSP9X mRNA were detected by RT-qPCR assay in HUVECs treated with RNase R or actinomycin D. D, F) Cell viability and circ_0090231 expression were assessed by MTT assay and RT-qPCR assay in HUVECs treated with ox-LDL at different concentrations (0 μg/ml, 25 μg/ml, 50 μg/ml, and 75 μg/ml) for 48 h. E, G) MTT assay and RT-qPCR assay were used to detect cell viability and circ_0090231 expression in HUVECs stimulated with 75 μg/ml ox-LDL for 0, 12, 24, 48, and 72 h. *p < 0.05
Deficiency of circ_0090231 blocked theangiogenesis and boosted oxidative stress, inflammatory response and apoptosis of ox-LDL induced HUVECs
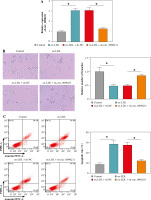
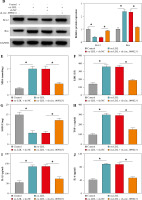
Next, to further explore the regulatory role of circ_0090231 in ox-LDL-caused HUVECs injury, we knocked down circ_0090231 expression in HUVECs. RT-qPCR assay revealed that the introduction of sh-circ_0090231 could counteract the promotion of ox-LDL on circ_0090231 expression in HUVECs (Fig. 2A). After that, tube formation assay showed that the treatment of ox-LDL retarded the ability of HUVECs to form a capillary-like structure, which was strikingly abolished by circ_0090231 downregulation (Fig. 2B). Meanwhile, the increased apoptosis rate caused by ox-LDL was effectively overturned by circ_0090231 silencing in HUVECs (Fig. 2C), accompanied by higher Bcl-2 (an anti-apoptosis factor) and lower Bax (a pro-apoptosis factor) (Fig. 2D). Apart from that, we further verified that the knockdown of circ_0090231 could mitigate ox-LDL-caused oxidative stress, exhibited as ox-LDL-mediated enhancement of MDA products and LDH activity, and decrease in SOD activity was abrogated by circ_0090231 deletion in HUVECs (Fig. 2E-G). In terms of inflammatory response, secretions of pro-inflammatory cytokines TNF-α, IL-1β, and IL-6 were significantly elevated in the presence of ox-LDL in HUVECs, which was weakened after the introduction of sh-circ_0090231 (Fig. 2H-J). Taken together, these data indicated that the downregulation of circ_0090231 could attenuate ox-LDL-induced HUVEC injury in vitro.
Fig. 2
Knockdown of circ_0090231 inhibited ox-LDL-mediated angiogenesis, apoptosis, oxidative stress, and inflammatory response in HUVECs. HUVECs were treated with control, ox-LDL, ox-LDL + sh-NC, and ox-LDL + shcirc_ 0090231. A) RT-qPCR assay was used to determine circ_0090231 level in treated HUVECs. B) Tube formation assay was applied to assess the number of capillary-like structures of HUVECs. C) Flow cytometry assay was performed to measure the apoptosis rate in treated HUVECs. *p < 0.05 D) Western blot assay was conducted to test the protein levels of Bcl-2 and Bax in treated HUVECs. E-G) The activities of MDA, LDH, and SOD were detected by commercially available kits in treated HUVECs. H-J) ELISA kits were used to examine the secretions of TNF-α, IL-1β, and IL-6 in treated HUVECs. *p < 0.05
Circ_0090231 could directly interact with miR-9-5p in HUVECs
In order to uncover the molecular mechanism of circ_0090231, the putative circ_0090231-interacting miRNAs were searched by the online software starBase. As shown in Figure 3A, miR-9-5p was found to have some complementary bases pairing with circ_0090231. The next dual-luciferase reporter assay revealed that the miR-9-5p elevation led to an overt reduction in the luciferase activity of circ_0090231 WT reporter vector but not that of circ_0090231 MUT in HUVECs (Fig. 3B). The interaction was further confirmed by RIP assay using Ago2, which was the core component of the RNA-induced silencing complex (RISC) [18]. Data showed that the enrichment of circ_0090231 was conspicuously improved by miR-9-5p upregulation using anti-Ago2 antibody in HUVECs (Fig. 3C), indicating the endogenous interaction between circ_0090231 and miR-9-5p. Moreover, the re-introduction of oe-circ_0090231 markedly promoted the facilitation of ox-LDL on circ_0090231 level in HUVECs (Fig. 3D). After that, we further identified the effect of circ_0090231 on miR-9-5p expression in ox-LDL-treated HUVECs. As displayed in Figure 3E, the deficiency of circ_0090231 could apparently augment miR-9-5p expression, while circ_0090231 overexpression could evidently block miR-9-5p in ox-LDL-exposed HUVECs. In a word, circ_0090231 modulates the abundance of miR-9-5p via binding to miR-9-5p.
Fig. 3
Circ_0090231 sequestered miR-9-5p in HUVECs. A) The biology website starBase predicted that there was a binding site between circ_0090231 and miR-9-5p. B) The binding was verified by dual-luciferase reporter assay. C) RIP assay was used to detect the enrichment of circ_0090231 in the RISC of HUVECs transfected with miR-NC mimic or miR-9-5p mimic using anti-Ago2 or IgG antibody. D) Circ_0090231 level was determined in HUVECs treated with control, ox-LDL, ox-LDL + vector, and ox-LDL + oe-circ_0090231 by RT-qPCR assay. E) miR-9-5p level was measured in HUVECs treated with control, ox-LDL, ox-LDL + sh-NC, ox-LDL + sh-circ_0090231, ox-LDL + vector, and ox-LDL + oecirc_ 0090231 by RT-qPCR assay. *p < 0.05
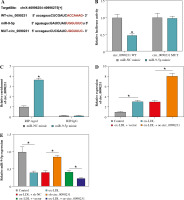
Downregulation of circ_0090231 could reverse ox-LDL-triggered HUVEC damage by targeting miR-9-5p
Considering the regulatory role of circ_0090231 in miR-9-5p expression in ox-LDL-treated HUVECs, we further explored whether the impact of circ_0090231 on ox-LDL- induced cell injury was associated with miR-9-5p. First of all, the transfection efficiency of miR-9-5p inhibitor in ox-LDL-triggered HUVECs was detected and presented in Figu- re 4A. Then, tube formation assay showed that circ_0090231 knockdown intensified the tube formation ability of HUVECs under the treatment of ox-LDL, which was significantly attenuated by miR-9-5p downregulation (Fig. 4B). Furthermore, the reduced expression of miR-9-5p could partly mitigate the suppressive action of sh-circ_0090231 on apoptosis in ox-LDL-treated HUVECs (Fig. 4C), as evidenced by decreased Bcl-2 and increased Bax (Fig. 4D). In addition, our results indicated that the negative effect of circ_0090231 downregulation on oxidative stress was effectively abrogated by the co-transfection of miR-9-5p inhibitor in ox-LDL-treated HUVECs, as described by improved MDA products and LDH activity, and reduced SOD activity (Fig. 4E-G). Consistently, ELISA assay demonstrated that the pro-inflammatory cytokines TNF-α, IL-1β, and IL-6 were reduced due to the silencing of circ_0090231, whereas miR-9-5p knockdown could distinctly relieve the effects in ox-LDL-induced HUVECs (Fig. 4H-J). All in all, these data implied that the downregulation of miR-9-5p could partly overturn the inhibitory effects of circ_0090231 deficiency on ox-LDL-caused HUVEC damage.
Fig. 4
Downregulation of miR-9-5p abolished the effects of circ_0090231 silencing on angiogenesis, apoptosis, oxidative stress, and inflammatory response in ox-LDL-treated HUVECs. A) miR-9-5p level was determined by RT-qPCR assay in HUVECs treated with control, ox-LDL, ox-LDL + miR-NC inhibitor, and ox-LDL + miR-9-5p inhibitor. B-J) HUVECs were treated with control, ox-LDL, ox-LDL + sh-NC, ox-LDL + sh-circ_0090231, ox-LDL + sh-circ_0090231 + miR-NC inhibitor, and ox-LDL + sh-circ_0090231 + miR-9-5p inhibitor. B) Tube formation assay presented the formation of capillary-like structures. C) Apoptosis rate of HUVECs was assessed by flow cytometry assay. *p < 0.05 D) Protein levels of Bcl-2 and Bax in HUVECs were measured by western blot assay. E-G) Special assay kits identified of MDA, LDH, and SOD products in treated HUVECs. *p < 0.05 H-J) The secretions of TNF-α, IL-1β, and IL-6 in treated HUVECs were detected by ELISA kits. *p < 0.05
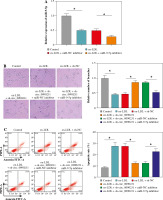
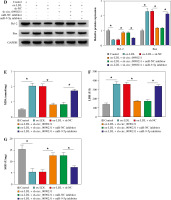
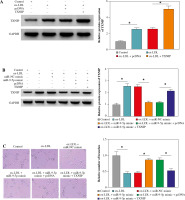
miR-9-5p directly interacted with TXNIP in HUVECs
To understand the mechanisms by which miR-9-5p mediates cell injury after ox-LDL treatment, we used starBase software to help identify the miR-9-5p target in HUVECs. As a result, there were some binding sites between miR-9-5p and TXNIP 3'UTR (Fig. 5A). To confirm the binding, a dual-luciferase reporter assay was carried out in HUVECs. The data revealed that the forced expression of miR-9-5p reduced the luciferase activity in the WT-TXNIP 3'UTR group compared with the MUT-TXNIP 3'UTR group (Fig. 5B), suggesting the binding relationship. Synchronously, the overexpression efficiency of miR-9-5p mimic in ox-LDL-induced HUVECs was determined and presented in Figure 5C. After that, we further assessed the authentic effect of miR-9-5p on TXNIP expression in ox-LDL-treated HUVECs. The data revealed that that the TXNIP protein level was decreased by exogenous miR-9-5p expression and reinforced by miR-9-5p knockdown in ox-LDL-triggered HUVECs compared with their respective controls (Fig. 5D). Of interest, our results showed that the downregulation of circ_0090231 could lead to an apparent reduction in TXNIP protein level, while the re-introduction of miR-9-5p inhibitor counteracted the effect in ox-LDL-exposed HUVECs (Fig. 5E), suggesting the regulatory role of circ_0090231/miR-9-5p/TXNIP axis in ox-LDL-challenged HUVECs. Collectively, these results suggest that TXNIP seems to be a key downstream effector of miR-9-5p.
Fig. 5
TXNIP was a target of miR-9-5p. A) The binding sequence between WT-TXNIP 3'UTR and miR-9-5p and the sequence of MUT-TXNIP 3'UTR. B) A dual-luciferase reporter assay was used to verify the binding. C) miR-9-5p level was determined by RT-qPCR assay in HUVECs treated with control, ox-LDL, ox-LDL + miR-NC mimic, and ox-LDL + miR-9-5p mimic. D) TXNIP protein level was measured by western blot assay in HUVECs treated with control, ox- LDL, ox-LDL + miR-NC mimic, ox-LDL + miR-9-5p mimic, ox-LDL + miR-NC inhibitor, and ox-LDL + miR-9-5p inhibitor. E) TXNIP protein level was assessed by western blot assay in HUVECs treated with control, ox-LDL, ox-LDL + sh-NC, ox-LDL + sh-circ_0090231, ox-LDL + sh-circ_0090231 + miR-NC inhibitor, and ox-LDL + sh-circ_0090231 + miR-9-5p inhibitor. *p < 0.05
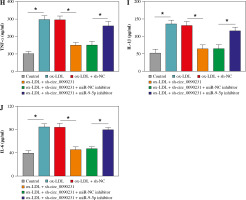
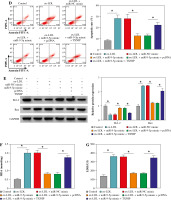
Overexpression of TXNIP partially overturned the repression of miR-9-5p on ox-LDL-caused HUVEC injury
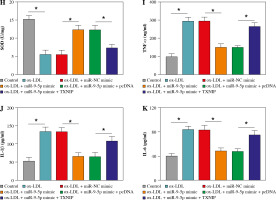
Then, to provide further mechanistic insight into the link between miR-9-5p and TXNIP in ox-LDL-triggered cell damage, we performed rescue assays. First, TXNIP was overexpressed in ox-LDL-treated HUVECs by transfection with pcDNA-TXNIP (Fig. 6A), and the transfection efficiency was confirmed to be successful. Subsequently, the results of western blot assay indicated that the co-transfection of TXNIP could notably ameliorate miR-9-5p upregulation-triggered enhancement in TXNIP protein level in ox-LDL-treated HUVECs (Fig. 6B). Functionally, miR-9-5p accumulation could increase the angiogenesis capacity of HUVECs after the treatment of ox-LDL, while the re-introduction of TXNIP abated the effect (Fig. 6C). Moreover, the suppression action of cell apoptosis on account of miR-9-5p upregulation in ox-LDL-treated HUVECs was recovered by TXNIP overexpression (Fig. 6D), as depicted by reduced Bcl-2 and augmented Bax (Fig. 6E). Apart from that, the forced expression of TXNIP could reverse the negative effect of miR-9-5p mimic on oxidative stress in ox-LDL-treated HUVECs, as indicated by higher MDA and LDH and lower SOD (Fig. 6F-H). Simultaneously, the pro-inflammatory cytokines, including TNF-α, IL-1β, and IL-6, were dampened, caused by miR-9-5p upregulation, which was partly reverted by TXNIP elevation in ox-LDL-induced HUVECs (Fig. 6I-K). All of these data suggested that miR-9-5p could attenuate ox-LDL-induced HUVEC damage by interacting with TXNIP.
Fig. 6
Upregulation of TXNIP partly overturned miR-9-5p-mediated angiogenesis, apoptosis, oxidative stress, and inflammatory response in ox-LDL-treated HUVECs. A) TXNIP protein level was examined by western blot assay in HUVECs treated with control, ox-LDL, ox-LDL + pcDNA, and ox-LDL + TXNIP. B-K) HUVECs were treated with control, ox-LDL, ox-LDL + miR-NC mimic, ox-LDL + miR-9-5p mimic, ox-LDL + miR-9-5p mimic + pcDNA, and ox-LDL + miR-9-5p mimic+TXNIP. B) TXNIP protein level was determined by western blot assay in treated HUVECs. C) Tube formation and apoptosis rate of HUVECs were detected by tube formation assay and flow cytometry assay. D) Tube formation and apoptosis rate of HUVECs were detected by tube formation assay and flow cytometry assay. E) Bcl-2 and Bax of HUVECs was measured by western blot assay. F, G) MDA, LDH products of treated HUVECs were analyzed by special assay kits. *p < 0.05 H) SOD products of treated HUVECs were analyzed by special assay kits. I-K) The secretions of TNF-α, IL-1β, and IL-6 of HUVECs were detected by ELISA kits. *p < 0.05
Discussion
With the development of high-throughput sequencing and bioinformatics analysis, more and more circRNAs have been identified in recent years [19, 20]. Due to being highly stable and abundantly expressed, circRNAs have been verified to be potential attractive biomarkers for human diseases [21, 22]. In practice, overwhelming evidence has indicated that the dysregulation of circRNAs has an inextricable correlation with the development of AS [23-25]. In this study, we found that circ_0090231 is a circular RNA from exons 3-12 of the USP9X gene, which was abnormally upregulated in ox-LDL-induced HUVECs, consistent with the former work [13]. Apart from that, ox-LDL-triggered HUVEC damage was established to simulate the pathogenesis of AS in this study.
Previous studies discovered that endothelial cell injury, such as abnormal angiogenesis, oxidative stress, and apoptosis, is responsible for breakdown of the integrity of the endothelial barrier, thus boosting the initiation and development of AS [26-29]. In this study, our results showed that the deficiency of circ_0090231 could abolish ox-LDL-caused decrease in angiogenesis oxidative stress, and enhancement in apoptosis in HUVECs. In addition, it was confirmed that inhibiting inflammation has a vital role for curbing the progression of AS [30, 31]. The present work suggested that the ox-LDL-induced inflammatory response in HUVECs was impeded by circ_0090231 downregulation. These data indicated that circ_0090231 silencing could repress ox-LDL-triggered endothelial cell damage and might be an underlying biomarker for AS treatment.
Recently, the circRNA-miRNA-mRNA regulatory mechanism has received increasing attention in various diseases, including AS [32, 33]. In this study, we first identified miR-9-5p, a latent circ_0090231-interacting miRNA using the starBase database. Apart from that, miR-9-5p has been documented to exert an inhibitory effect in AS through repressing vulnerable plaque formation and vascular remodeling [16, 17]. Furthermore, miR-9-5p was previously confirmed to attenuate AS-associated inflammation by JAK1/STAT signaling [34]. In the present study, the downregulation of miR-9-5p in ox-LDL-exposed HUVECs was observed for the first time. Also, the functional analysis underscored that the miR-9-5p reduction could relieve the suppressive action of circ_0090231 knockdown on ox-LDL-caused endothelial cell injury in vitro. Likewise, TXNIP as the potential target mRNA of miR-9-5p, TXNIP was acquired using the online software in this research. In agreement with the prior study, TXNIP expression was increased in ox-LDL-stimulated HUVECs [35]. Also, TXNIP is an inhibitor of thioredoxin, which could block neovascularization, and induce apoptosis and inflammation [36-38]. The current work indicated that miR-9-5p could restrain endothelial cell dysfunction through downregulating TXNIP in ox-LDL-triggered HUVECs. The promotion of TXNIP on ox-LDL-caused endothelial cell damage was also proved in a previous report [12]. Additionally, miR-9-5p knockdown partly reversed circ_0090231 deficiency-mediated decline of TXNIP expression, further suggesting that the circ_0090231-miR-9-5p-TXNIP regulatory network might partake in AS progression.
Conclusions
Taken together, these results demonstrated that circ_0090231 could regulate ox-LDL-triggered cell damage in HUVECs in part by targeting the miR-9-5p/TXNIP axis (Fig. 7), providing a crucial preclinical basis for AS treatment.