Introduction
Human monocytes, macrophages and dendritic cells (DCs) constitute a compartment that initiates the first line of defense against microbial invasion by the regulation of innate and adaptive immune responses [1, 2]. The mobilization and differentiation from monocytes to immature dendritic cells (iDCs) is one of the key processes involved in the homeostasis of the immune system. A particular stimulus or a combination such as interleukin (IL)-6, interferon γ (IFN-γ), granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-4 or microbial pattern-associated agents can drive the differentiation of iDCs to a particular subset of DC which enhances or suppresses the immune response [3, 4]. Mature DCs are fully activated DCs that deliver activating signals to T cells by the production of IL-12, IL-6, and IFN-γ and the expression of co-stimulating molecules such as CD86 and CD80. In contrast, tolerogenic DCs could also be induced from immature DCs to negatively regulate the immune response by induction of regulatory T cells and induction of T cell anergy, production of immunosuppressive molecules such as IL-10, IL-27, nitric oxide, etc. [1, 5].
MicroRNAs (miRNAs) are highly conserved, small non-coding RNA molecules containing approximately 22 nucleotides that regulate gene expression by binding to 3’-UTR regions of mRNA. These molecules are essential in cell differentiations, cell-cell interactions and cellular responses to stimuli. In the immune system, miRNAs play crucial roles in regulating the activation and differentiation of immune cells and consequently modulate cytokine release, antibody production, etc. [6, 7]. Previous studies showed that miRNAs regulate the functions of DCs through signaling mediators such as p27kip1, KPC1, and SOCS-1, or tumor-derived exosomes [8, 9]. MiR-155 and miR-142 were reported to be involved in the modulation of DC maturation [8-10]. Analysis of individual miRNAs in terms of immune cells was studied by previous research groups [6-14]; however, overall analyses of miRNAs using microarray could display the comprehensive expression pattern of miRNA and facilitate further investigation on the regulation of immune responses by miRNA.
This study aimed to depict the profile of miRNA in human monocytes and iDCs and to identify the key miRNAs that are involved in the differentiation of iDCs. Human monocytes were cultured in vitro and induced by IL-4 plus GM-CSF to differentiate into DCs. MiRNA microarray chips were utilized to analyze the changes of the expression of miRNAs in both monocytes and iDCs. The results showed that miR-122a and miR200c exhibited the most striking changes involved in the differentiation of iDCs. The results of this study help understand the regulation of iDC development by miRNA.
Material and methods
Study participants
The peripheral blood samples were collected from healthy volunteers (n = 5) recruited in Tianjin Medical University General Hospital. Written, informed consent was obtained from all participating individuals before the procedure. The study was approved by the Ethical Committee of Tianjin Medical University General Hospital.
Differentiation of monocytes into iDCs
For iDC generation, monocytes were isolated by adhesion from Ficoll gradient-separated peripheral blood mononuclear cells (PBMCs). The cell preparation was tested for purity by flow cytometry. Monocytes were identified as CD14(+) HLA-DR(+) cells. The purity from this purification was in excess of 90%. Monocytes were subjected to cytokine-driven differentiation into iDCs by 6-day culture in the presence of GM-CSF and IL-4, and iDC cell differentiation was evaluated by flow cytometry on day 6. Differentiated iDCs were defined as DCSIGN(+) CD1a(+) CD206(+) HLA-DR(high) CD86(high) CD14(–) and CD15(–) cells. iDC yields following culture were > 95%.
Microarray profiling of miRNAs
Microarray analysis of 856 miRNAs was carried out by µParaflo technology (LC Sciences, TX, USA). Data signals were collected by an Axon GenePix 4000B Microarray Scanner. A total of 856 human mature miRNA are listed in the Sanger miRNA Sequence Database (miRBase Release 12.0). Total RNAs were extracted from the adherent monocytic fraction (107 cells) or cytokine differentiated iDCs (n = 5) using a mirVana miRNA Isolation kit (Ambion, TX, USA) according to the manufacturer’s instructions. The miRNA samples were label-blinded and subjected to array hybridization. The miRNAs that differed quantitatively (at least 2-fold) from counterpart monocytes in differential subtraction analysis were considered to be remarkably changed miRNAs illustrating that DC differentiation was accompanied by definable changes in miRNA expression. miRNAs that displayed profound differential expression (at least 4-fold) were selected for confirmatory analysis by real-time PCR analysis using pre-validated miRNA-specific primers (Applied Biosystems, Foster City, CA).
Real-time PCR analysis
MiRNAs were extracted using a mirVana miRNA Isolation kit (Ambion, TX, USA). cDNAs were synthesized by a miRScript II RT kit (Qiagen) according to the manufacturer’s instructions. The expression of miRNA was determined using the TaqMan MicroRNA assays (Applied Biosystems) according to the manufacturer’s protocol. miRNA expression was normalized to RNU43, a small nuclear RNA endogenous control. Expression levels of the following miRNAs were assayed: miR200c, miR335, miR514, miR509, miR31, miR442b, miR1, miR199a, miR203, miR363, miR489 miR122a, miR210, miR155, miR126, miR139, miR452, miR19a, miR25 and miR181d. The expression data were analyzed using the 2-ΔΔCT method.
Statistical analysis
For microarray data analysis, the ANOVA test was used to evaluate the differences between monocytes and iDCs. When the differences of expression were log2 fold-change > 2 or log2 fold-change < –2, it is considered meaningful. Student’s t test was applied to compare the differences in the expression of miRNAs between the monocyte group and iDC group measured by real-time PCR. Statistical significance was set at two-tailed p < 0.05.
Results
Detectable miRNAs expressed in human monocytes and iDCs with miRNA microarray
Human monocytes were isolated from peripheral venous blood and cultured in the presence of IL-4 and GM-CSF to drive their differentiation to DCs. Cells from five donors were prepared and analyzed by miRNA microarray which contains 856 miRNAs spotted on the chip. The microarray detected a total of 381 miRNAs in which 152 miRNAs were present in both monocytes and iDCs. There were 3 miRNAs which were detectable only in monocytes whereas 74 miRNAs were present only in iDCs. There were 103 miRNAs differentially expressed between monocytes and iDCs (Fig. 1). The fluorescence signals representing miRNA expression were plotted as a heatmap and all detectable miRNAs were listed (Fig. 2). The results demonstrated that much more miRNAs were expressed in iDCs than in monocytes, implying that miRNA might play a more important role in keeping biological function of iDCs.
Fig. 1
Numbers of detectable miRNAs in the experimental groups analyzed by miRNA microarray. Human monocytes were differentiated to dendritic cells (DCs) in culture with IL-4 and GM-CSF. MiRNAs extracted from monocytes and iDCs (n = 5) were subject to microarray hybridization in a blinded manner. MiRNAs were considered detectable when both Cy3 and Cy5 signals were reported by the instrument. Differential expression was defined by at least twofold changes in signal intensity in iDCs when compared to that in monocyte control in differential subtraction analysis
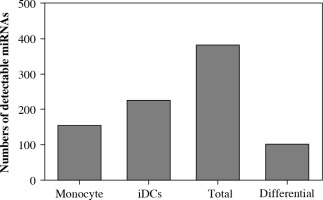
Fig. 2
Heatmap illustration of miRNAs expressed in iDCs and the original control monocytes. MiRNAs extracted from both monocytes and iDCs were subject to microarray hybridization containing 587 miRNAs spotted on the chip. Detectable miRNAs were plotted on a heatmap using Heml v1.0 software. Black color refers to undetectable signals which were recorded as 0 as digitized signals. Red color indicates the highest expression and blue color indicates the lowest expression

Differential expression of miRNAs between human monocytes and induced iDCs
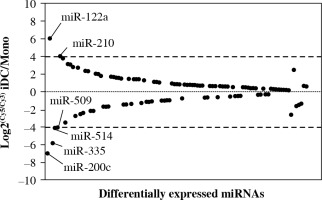
It is of interest to note that there were a number of profoundly altered miRNAs involved in the differentiation of iDCs from monocytes. One hundred and three miRNAs out of 381 detectable miRNAs in both monocytes and iDCs were differentially expressed miRNAs (Table 1). MiR-122a and miR-210 were markedly upregulated whereas miR-509, miR-514, miR-335 and miR-200c were downregulated at the cutoff line of 4-fold change (Fig. 3). There were 20 miRNAs that exhibited differential expression levels when the cutoff line was set at 2-fold change. In this regard, the upregulated miRNAs included miR-31, miR-442b, miR-1, miR-199a, miR-203, miR-363 and miR-489 whereas the upregulation of miR-155, miR-126, miR-139, miR-452, miR-19a, miR-25 and miR-181d was considerable. The results provided an overview of the miRNA profile implicated in the differentiation of iDCs from monocytes in humans.
Fig. 3
Differential expression of miRNAs in monocytes and derived iDCs. The differentially expressed miRNAs were defined as at least 2-fold for log2(Cy5/Cy3) for the ratio of expression in iDCs to monocytes. The Y-axis indicates the ratio of miRNA expression. The X-axis indicates each individual of differentially expressed miRNA. When the ratio was beyond 4-fold changes, the names of those miRNA were labeled in the plot. These miRNAs were selected for confirmatory analyses using real-time PCR
Analysis of expression of miRNAs by real-time PCR
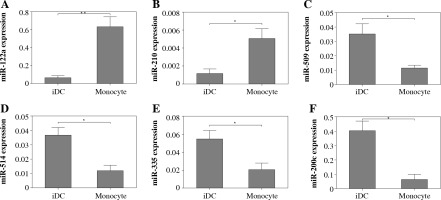
To confirm the expression of those miRNA with pronounced difference in monocytes and iDCs, we utilized real-time PCR to determine the expression of six individual miRNAs selected with at least 4-fold alteration between monocytes and differentiated DCs. The expression of both miR-122a and miR-210 was significantly elevated in iDCs as compared to monocytes. In contrast, the expression levels of miR-509, miR-514, miR-335 and miR-200c were found to be decreased significantly in iDCs as compared to monocytes (Fig. 4). The results of real-time PCR were consistent with the data generated by miRNA microarray.
Fig. 4
Real-time PCR analysis of the expression of selected miRNAs. The expression of selected miRNAs was determined using real-time PCR. MiRNA expression was normalized to RNU43 small nuclear RNA endogenous controls. The relative expression of miRNA was calculated using the 2–ΔΔCT method. The data were expressed as mean ± standard deviation of multiplicate wells. Asterisks refer to p < 0.05 calculated using Student’s t test
Discussion
The miRNA system is a complex network that largely silences or degrades the transcripts of mRNA by being incorporated into Argonaute (Ago) protein within the RNA-induced silencing complex [15-21]. In developmental biology, miRNAs have long been known to be involved in cell differentiation and organ development [22]. Previous efforts have been made to understand the regulation of immune cells by miRNA. In T cell and B cell developmental and differentiation, several miRNAs were reported to play important roles in the differentiation of T cells such as miR-150, miR155, miR-181, miR-17-92; B cells such as miR-101, miR-152, miR-155, miR-181, miR-17-92; and Tregs such as miR-146, miR-150, miR1-55 [23-36]. In the innate immune system, miRNA contributes to the maturation of DCs. miR155 was induced by activating the NF-kB and Jun kinase pathway and targeted SOCS1 to regulate the maturation of DCs [7]. miR146a was involved in the TLR signaling cascade in DC maturation [37]. miR155 was considered to be an important miRNA that regulates the immune cells as evident by these previous reports. In our study, the data showed that miR155 was also markedly upregulated in iDC (the value of Log2(Cy5/Cy3) was 3.77), as seen in Table 1. In addition to the known miRNAs reported elsewhere, our results revealed that miR122a was most strikingly upregulated in iDCs derived from human monocytes upon GM-CSF and IL-4 stimulation in vitro.
Table 1
Differential expression of miRNAs in iDCs derived from human monocytes
In this study, we found that the expression of miR200c was the lowest among all decreased miRNAs in iDCs. miR200c was reported to be associated with the growth of tumors [38, 39]. Another downregulated miRNA, miR335, was also found to be related to the pathogenesis of bladder, prostate and breast cancers [40-42]. The downstream targets of miR335 could be multiple events such as ROCK1, NO synthase, etc. The results revealed several important miRNAs implicated in the cytokine-induced differentiation from monocytes to iDCs.



