Introduction
Coronavirus disease 2019 (COVID-19) is an acute respiratory infection caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) [1]. Around 81% of COVID-19 patients have moderate symptoms and recover without supportive care, and 14% of patients suffer from severe respiratory distress, though roughly 5% are classified as critical cases owing to fast respiratory failure necessitating ventilatory support and specialist treatment in intensive care units (ICU) [2]. As of April 21, 2024, 775,364,261 confirmed cases and 7,046,320 confirmed deaths had been reported by the World Health Organization (WHO). Thus, about 0.9% of infected people died after infection with this virus [3].
Weakening of the integrity of the host surface or weakened host immunity can cause fungal infections that can affect organs and lead to catastrophic illnesses such as aspergillosis or fungal pneumonia. Fungal infections can manifest as superficial, deep, or systemic conditions [4]. COVID-19 increases the risk of fungal pathogen co-infections in the elderly and immunocompromised patients. Candidiasis, aspergillosis, and mucormycosis are the most frequent fungal pathogen infections seen in people with COVID-19 [5, 6].
Monocytes and neutrophils are triggered by pathogen-associated molecular patterns (PAMPs) via specialized receptors located on their surface, known as pattern recognition receptors (PRRs) [7, 8]. Dectin-1 is also known as C-type lectin domain family member A (CLEC7A), serving as a prominent PRR. It is largely expressed by neutrophils, monocytes, dendritic cells and macrophages [9, 10]. Dectin-1 is critical for pathogen detection and signaling in innate immunity [11, 12], particularly against fungal pathogens by detecting fungal β-glucans, which prompt the immune cell to engulf the pathogens and release essential molecules necessary for eliminating them [13]. Additionally, dectin-1 directly influences the adaptive immune response, particularly affecting the T-helper 1 (Th1) and Th17 pathways [14, 15]. Dectin-1 can identify β-glucans found in fungi, which leads to the generation of soluble mediators and phagocytosis to eliminate the fungus. Through Th1 and Th17, dectin-1 can also influence the adaptive immune system. Thus, dectin-1 and its coding gene impact innate and adaptive immune systems [16, 17].
Toll-like receptor 2 (TLR2), a PRR distributed across diverse immune cells, also plays a vital role in activating both innate and adaptive immune responses and plays a crucial part in the immune system’s defense mechanism against microbial invasion by recognizing certain molecular patterns on a microorganism’s surface [18, 19]. Human PRR polymorphisms have been correlated with an elevated susceptibility to infectious complications. A functional single nucleotide polymorphism (SNP) in the dectin-1 gene known as Y238X (rs16910526) introduces an early termination codon resulting in the truncation of the last 10 amino acids of the carbohydrate-recognition domain of the dectin-1 receptor, leading to reduced surface expression on immune cells [20, 21]. Polymorphisms of TLR2 and dectin-1 also increase susceptibility to fungal infection in non-COVID-19 patients, according to a study conducted by Fischer et al. [21], Polymorphisms of dectin-1 (rs7309123) and TLR2 (rs5743708) are independent risk factors for systemic fungal infection in acute myeloid leukemia (AML) patients receiving induction chemotherapy. Additionally, functionally significant mutations within the TLR2 gene, such as Arg753Gin (rs5743708), significantly raise the risk of infectious complications such as pneumonia and sepsis [21, 22]. Increased Candida colonization in hematopoietic stem cell transplantation (HSCT) recipients has been linked to the Y238X polymorphism in dectin-1, which suggests compromised mucosal immunity [23]. In their systematic review, Aghaei et al. [24] discovered a link between dectin-1 rs16910526 and hematological malignancies such as AML, lymphoma, and multiple myeloma.
Regarding SARS-CoV-2 infection and gene polymorphism, according to the research, the presence of the TLR2 rs5743708 mutant (G/A) genotype and A allele was significantly associated with a higher risk of SARS-CoV-2 infection and served as a major risk factor for developing pneumonia in COVID-19 [25, 26]. Furthermore, the risk of COVID-19 occurrence is associated with TLR2 rs5743708 variants and is linked to more severe symptoms and poor prognosis in Egyptian patients [27]. Currently, we are unaware of any findings suggesting a correlation between the dectin-1 rs16910526 variant and susceptibility to SARS-CoV-2 or fungal infections in COVID-19. Although there is no direct evidence linking rs16910526 to COVID-19, the presence of dectin-1 variants could potentially affect immune responses, leading to variations in disease severity and susceptibility to fungal infection.
During infections and inflammatory conditions, soluble TLRs are released into the circulation from tissues and blood cells, causing an increase in their concentrations [28]. Few studies have measured the serum levels of dectin-1, as it is mainly expressed in tissues. Kalia et al. [29] aimed to determine the serum level of dectin-1 and its association with the genotype (rs3901533) in recurrent vulvovaginal fungal infection.
The aim of this investigation was to detect the potential impact of the TLR2 SNP rs5743708 and the dectin-1 SNP rs16910526 on the sensitivity to fungal infection in patients diagnosed with COVID-19 and ultimately providing insights into personalized risk assessment and targeted therapeutic interventions for this vulnerable population. Additionally, the study sought to determine the functional association between the TLR2 SNP rs5743708 and dectin-1 SNP rs16910526 and serum levels of TLR2 and dectin-1.
Material and methods
Patients and study samples
This prospective multi-center observational study took place from May 1, 2021 to October 1, 2021, with patients who attended ICU departments of the hospitals in the Kurdistan region of Iraq, comprising four cities (Erbil, Duhok, Sulaymaniyah, and Kirkuk). In all, 187 non-vaccinated COVID-19 patients were included in the study and were classified into two groups: COVID-19 free from fungal infection (COVID-19 FFI) (n = 110) and COVID-19 with fungal infection (COVID-19 WFI) (n = 77). The sample size was calculated based on the G*Power program [30], according to the following formula [31]:
This equation was simplified by combining unequal sample size groups:
where:
n2 = sample size for COVID-19 WFI,
r = 1.43 (ratio of COVID-19 FFI to COVID-19 WFI),
Zα/2 = 1.96 (for a significance level of 0.05, two-tailed),
Zβ = 1.645 (for 95% power),
d = 0.6406 is the effect size
The modified equation was used to calculate the sample size for the larger group (n1):
n1 = r * n2
where:
n1 = sample size for COVID-19 FFI
r = 1.43
n 2 = 77
n 1 = 1.43 * 77 = 110
Following WHO guidelines, the COVID-19-positive patients were verified using molecular testing (RT-PCR), laboratory testing, and radiographic findings [32]. Fungal infection was confirmed in a fungal laboratory via culture methods. Blood samples and endotracheal aspirate (ETA) were collected from all participants. The blood was placed in an EDTA tube for genetic analysis and a serum separation tube for serological tests. It was stored at –80°C until needed.
The medical history, comorbidities, and demographic information were gathered using the hospital’s data management system. This project was authorized by Salahaddin University-Erbil’s (SUE) Ethics Committee (approval number: R03-021; 92 on April 15, 2021), and before blood sampling, all subjects provided written informed consent. We questioned the relatives of patients who were in a critical condition and could not sign; these individuals served as the patient’s replacement decision-makers.
The inclusion criteria for COVID-19 patients were to be non-vaccinated, positive for SARS-CoV-2 by RT-PCR, having complete demographic data and samples (blood and nasopharyngeal), having signed a consent form, and being of Kurdish ethnicity.
Human blood DNA extraction
The deoxyribonucleic acid (DNA) extraction was processed in compliance with the manufacturer’s protocol (Jena Bioscience, Germany). In the study, 300 µl of each sample of whole blood was placed into 1.5 ml micro-centrifuge tubes (Citotest, Haimen, China). Then, 900 µl of red blood cell lysis buffer was pipetted into the tubes and thoroughly shaken before being left at room temperature for 3 minutes, with occasional inversion. The tubes were then centrifuged for 30 seconds at 13,000 rpm, and the fluid was removed using a pipette, leaving the discernible cell pellet. The white cells in the residual liquid were mixed back together by vigorously shaking the tubes for 10 seconds. 300 µl of cell lysis solution was put into the re-suspended cells and pipetted up and down to break down the cells until no clumps could be seen.
In order to precipitate the proteins, the cell lysate was agitated vigorously for 20 seconds with 100 µl of protein precipitation solution to ensure that tiny particles of precipitated protein (without clumps) were visible. After that, the proteins precipitated, and a tight, dark pellet was formed when the tubes were centrifuged at 13,000 rpm for 1 minute. We added 300 µl of isopropanol > 99% (BDH Chemicals, Poole, UK) to a clean 1.5 ml micro-centrifuge tube (Citotest, Haimen, China) to precipitate the DNA. Then we combined the supernatant and mixed the samples by gently inverting for 1 minute and then centrifuging at 13,000 rpm for 1 minute. Finally, the extracted DNA samples were stored at –80°C for downstream applications.
DNA concentration and purity determination
The concentration of DNA of the samples was measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, United States) with the unit ng/µl. At 260 and 280 nm, optical density was measured. DNA purity was assessed using the ratio of absorbance at 260/280 nm. This work was done in the Bio-lab, Erbil, Iraq. The probe of the NanoDrop spectrophotometer was cleaned with deionized H2O (diH2O). The NanoDrop was blanked by adding 2 µl of diH2O and then Elution buffer. The probe of NanoDrop was cleaned again, and DNA was measured by adding 2 µl of DNA samples. The range of concentration and purity of DNA was 13.4-126 and 1.62-1.93 ng/µl, respectively.
Genotyping of SNPs in TLR2 (rs5743708) and dectin-1 (rs16910526)
Sanger sequencing and traditional PCR were used to examine the SNP in the TLR2 and dectin-1 genes. The primers were designed by the GeneRunner program and checked using NCBI primer blast. Table 1 displays the primers and SNPs of the investigated genes in this study. These primers were ordered from Metabion Company (Germany). PCR materials contained 2 µl of genomic DNA, 10 µl of Mastermix (Ampliqon, Denmark), 1 µl of each primer, and 4 µl of diH2O in a 20 µl reaction tube. The thermocycling protocol began with an initial denaturation step at 94°C for seven minutes, followed by 30 cycles of 94°C for 40 seconds, 57°C for 40 seconds, 72°C for one minute, and a final extension of 72°C for five minutes.
Table 1
Genes, single nucleotide polymorphisms (SNPs) and sequence of primers involved in this study
| Primer sequences 5′-3′ | Band size | ||
|---|---|---|---|
| TLR2 (rs5743708) | |||
| Forward | GAAAGCTCCCAGCAGGAACA | 582 bp | |
| Reverse | AGCAAGTCCTCAAATGACGGT | ||
| Dectin-1 (rs16910526) | |||
| Forward | AGAGTTCTCCCTGAGGTCTGAA | 948 bp | |
| Reverse | TTCTCTCTCCTTCTCCACCCTT | ||
Following the addition of the PCR products, a 2% agarose gel electrophoresis stained with ethidium bromide was visible under a UV lamp. All samples’ PCR results were forwarded for Sanger sequencing once the intended band was confirmed in the gel (Zheen International Hospital, Iraq). It was analyzed via the Geneious prime program and mutation surveyor and confirmed by NCBI nucleotide blast. Two Sanger sequences were submitted to NCBI and received accession codes (PQ140773 for TLR2 and PQ140774 for dectin-1).
Serum TLR2 and dectin-1 assays
Serum samples from all COVID-19 patients were analyzed using ELISAs to assess the amounts of TLR2 and dectin-1. ELISA kits from RayBiotech, USA were employed for our experiments. We used the human TLR2 ELISA kit (accession number O60603) with a detection range of 0.32 ng/ml to 80 ng/ml, and the Dectin-1 ELISA kit (accession number Q9BXN2) with a detection range of 20 ng/ml to 0.082 ng/ml, following the manufacturer’s instructions.
Fungal isolation
Special pulmonologist physicians collected the ETA in appropriate containers, following the CDC’s standard protocol for specimen collection, transportation, and processing, as discussed by Vianna et al. [33]. Specimens were transferred to the microbiological laboratory and subsequently transferred to the Mycological Laboratory in the College of Sciences, Department of Biology, Salahaddin University.
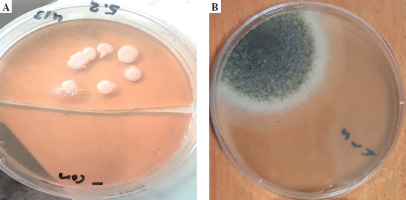
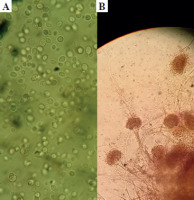
The ETA was initially inoculated onto Sabouraud dextrose agar medium (SDA), enriched with chloramphenicol, and incubated at 37°C. The presence of white, creamy colonies suggested Candida spp. Next, further research was conducted using the germ tube test, and differentiating between Candida spp. was done using CHROMagar Candida (CHROMagar, France). The mycological identification of mold was based on macroscopic (Fig. 1) and microscopic (Fig. 2) examination of the culture isolates. Aspergillus species were identified as recommended by Pitt & Hocking [34].
Statistical analysis
All the data were normally distributed and passed normality tests (D’Agostino, Kolmogorov-Smirnov and Shapiro-Wilk tests). Independent t-tests, one-way ANOVA and chi-square tests were utilized to analyze the clinical and demographic data. The statistical analysis was performed using SPSS Statistics 27 (IBM, USA), MedCalc20 (MedCalc Software Ltd., Belgium), and GraphPad 9 (GraphPad Software Inc., USA). The data were presented as mean ± standard error of mean (SEM). A p-value of < 0.05 was considered statistically significant.
Results
Demographic characteristics of study subjects
The total participants included in the study were 187 pa- tients (Table 2), 110 COVID-19 FFI and 77 COVID-19 WFI. The mean age of COVID-19 FFI was 41.55 ±1.236 years (mean±SEM) including 60 men and 50 women. The mean age of COVID-19 WFI was 40.24 ±0.911 years, including 44 men and 33 women. There was no significant difference in the age of COVID-19 FFI and COVID-19 WFI (p-value 0.431). Additionally, there were no significant differences in body mass index (BMI) between COVID-19 FFI (25.85 ±0.654) and COVID-19 FFI patients (mean of BMI = 25.76 ±0.431) with a p-value 0.916.
Table 2
Demographic characteristics of COVID-19 FFI and COVID-19 WFI
| Parameters | COVID-19 FFI Mean ±SEM | COVID-19 WFI Mean ±SEM | p-value (OR) |
|---|---|---|---|
| n | 110 | 77 | – |
| Gender (M/F) | 60/50 | 44/33 | 0.90 (1.111) |
| Age (years) | 41.55 ±1.236 | 40.24 ±0.911 | 0.431 |
| BMI (kg/m2) | 25.85 ±0.654 | 25.76 ±0.431 | 0.916 |
[i] Age and BMI were compared between groups using an independent t-test in SPSS27, while the association of gender with susceptibility of fungal infection was determined using the chi-square test in GraphPad 9. The data are presented as mean ± standard error of mean (SEM). Statistical significance was determined with a p-value less than 0.05.
Associations of TLR2 (rs5743708) and dectin-1 (rs16910526) SNPs with susceptibility to fungal infection in COVID-19
The presence of TLR2 (rs5743708) and dectin-1 (rs16910526) SNPs in COVID-19 FFI and COVID-19 WFI was examined (Fig. 3). Regarding the TLR2 SNP rs5743708, we found that the AG genotype had a similar association with fungal infection as the AA genotype (p = 0.99). On the other hand, the GG genotype and G allele were found to decrease the risk of being infected with fungal infection (p < 0.0001, odds ratio [OR] = 26, 95% CI: 8.506-71.43; and p < 0.0001, OR = 5.906, 95% CI: 3.704-9.302, respectively). In comparison to AA and GG genotype, the recessive model (AG + GG) and the dominant model (AA + AG) were both linked to fungal infection compared to the AA and GG genotypes (Table 3).
Fig. 3
Gel electrophoresis result of SNP in toll-like receptor 2 (TLR2) and dectin-1 PCR products. The 50 bp marker was used. S1 to S2 contain samples for dectin-1; the size for the PCR product is 948 bp. On the other hand, S3 to S4 contain samples for TLR2. The size for the PCR product is 582 bp. L50 – ladder 50 bp
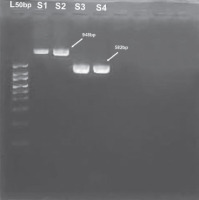
Table 3
Distribution of genotype and alleles of the TLR SNP (rs5743708) was compared between COVID-19 FFI and COVID-19 WFI
[i] The statistical analysis was conducted via chi-square (χ2) test using GraphPad Prism 9. Statistical significance was assumed if the p-value was < 0.05. COVID-19 – coronavirus disease 2019, CI – confidence interval, n – sample size, OR – odds ratio, COVID-19 FFI – COVID-19 patients free from fungal infection, COVID-19 WFI – COVID-19 patients with fungal infection
Regarding the dectin-1 SNP (rs16910526), we found no significant difference in the association with fungal infections between the TG and the TT genotypes (p = 0.99, OR = 1.056, 95% CI: 0.504-2.201). In contrast, the GG genotype and G allele were found to be associated with increased risk of being infected with fungal infection in COVID-19 (p < 0.0001, OR = 7.741, 95% CI: 3.347-17.69; and p < 0.0001, OR = 3.507, 95% CI: 2.277-5.421, respectively). Regarding the recessive model (TG + GG) and dominant model (TT + TG), both models of genotypes showed positive associations with fungal infection in COVID-19 when compared to the AA genotype and GG genotypes (Table 4).
Table 4
Distribution of genotype and alleles of the dectin-1 SNP (rs16910526) was compared between COVID-19 FFI and COVID-19 WFI cases
[i] The statistical analysis was conducted via chi-square (χ2) test using GraphPad Prism 9. Statistical significance was assumed if the p-value was < 0.05. COVID-19 – coronavirus disease 2019, CI – confidence interval, n – sample size, OR – odds ratio, COVID-19 FFI – COVID-19 patients free from fungal infection, COVID-19 WFI – COVID-19 patients with fungal infection.
Fungal culture isolated from COVID-19 patients
The fungal culture isolated from the specimens of COVID-19 included Candida spp. in 51 cases (27.272%), making it the most common organism, and Aspergillus spp. was found in 26 cases (13.903%), while there was no fungal growth in 110 cases (58.823%), as shown in Table 5.
Assessing the functionality of TLR2 and dectin-1
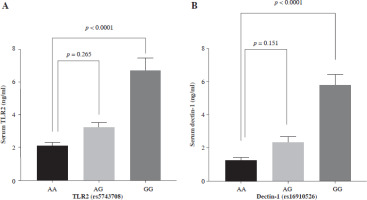
In Table 6, it can be observed that COVID-19 WFI had significantly higher levels of serum TLR2 and dectin-1 compared to COVID-19 FFI (p < 0.001). In all COVID-19 cases, serum TLR2 and dectin-1 levels were segregated based on different genotypes (Fig. 4A, B). The GG genotype of dectin-1 (rs16910526) had a significant impact (p < 0.0001) on serum dectin-1 levels (5.854 ±0.677), while the mean of serum dectin-1 produced by the TG genotype (2.376 ±0.3610) did not show a significant difference (p = 0.151) compared to the serum of the TT genotype (1.240 ±0.191) in all COVID-19 cases. Serum TLR2 levels were significantly higher (p < 0.0001) for GG genotypes (6.715 ±0.747) of TLR2 (rs5743708) compared to the AA genotype (2.089 ±0.205). There was no observed difference (p = 0.265) in genotypic serum TLR2 levels between AA (2.089 ±0.205) and AG genotypes (3.181 ±0.351) of TLR2 (rs5743708).
Table 6
Serum TLR2 and dectin-1 of COVID-19 FFI and COVID-19 WFI
| Parameters | COVID-19 FFI Mean ±SEM | COVID-19 WFI Mean ±SEM | p-value |
|---|---|---|---|
| Serum TLR2 (ng/ml) | 2.327 ±0.1688 | 6.646 ±0.6832 | < 0.0001 |
| Serum dectin-1 (ng/ml) | 1.408 ±0.1627 | 5.646 ±0.5876 | < 0.0001 |
Fig. 4
Serum toll-like receptor 2 (TLR2) and dectin-1 levels produced by different genotypes (A) serum TLR2 production by TT, TG and GG genotypes of TLR rs5743708 (B) serum dectin-1 production by AA, AG and GG genotypes of dectin-1 rs16910526. One-way ANOVA in GraphPad Prism was used to compare between groups, followed by multiple comparison using Dunnett’s test. Data are presented as mean ± standard error of the mean (SEM). Probability (p) value less than 0.05 was considered significant
Discussion
The emergence of fungal coinfections in COVID-19 patients has raised significant concerns regarding disease severity and mortality. Numerous predisposing variables, including genetic factors, have been shown in epidemiological studies to influence the likelihood of having FI. Understanding the genetic predispositions underlying susceptibility to fungal infections in COVID-19 is imperative for tailored therapeutics and improved patient outcomes [35]. The study’s main results were an association between fungal coinfections in COVID-19 and TLR2 (rs5743708) and dectin-1 (rs16910526) gene polymorphisms.
During the response to infections, innate immunity plays a major part in the immunopathogenesis of inflammatory reactions. Immune cells, fibroblasts, and epithelial cells, including type II pneumocytes, all express TLRs [36]. Studying the SNP allele in association with genes related to both acquired and innate immunity significantly impacts the entire course of influenza-related pneumonia infections [37]. Depending on our data analysis, the SNPs listed in Table 2 shed light on the potential impact of genetic variants in modulating sensitivity and susceptibility to fungal coinfections in COVID-19.
The fungal culture isolated from COVID-19 specimens comprised Candida spp. in 51 instances (27.272%), making it the most prevalent organism, and Aspergillus spp. in 26 cases (13.903%). Our results are in line with the research conducted by Negm et al. [38], who stated that the most often isolated fungus was Candida in 61 (24.1%) of 253 followed by Aspergillus in 11 (4.3%) in ICU COVID-19 patients.
Individuals carrying the GG genotype of TLR2 rs5743708 exhibited a substantially higher risk of fungal coinfections in COVID-19 than those with the AA genotype. Furthermore, the G allele of TLR2 rs5743708 was also significantly associated with increased susceptibility to fungal coinfections. These findings suggest that genetic variations in TLR2 may influence host immune responses to fungal pathogens during COVID-19, and these results were comparable to the study done by Alhabibi et al. [25], as they reported a significant association between TLR2 rs5743708 variants and an increased susceptibility to and severity of COVID-19 infection. Similarly, Alhabibi et al. [25] discovered that the A allele and the TLR2 rs5743708 mutant (G/A) genotype were substantially linked to an increased probability of contracting COVID-19 infection and were a significant risk factor for the severity of the infection.
The PRR dectin-1 can first identify fungal pathogens through PAMPs in the fungal cell walls. Then, an activation cascade starts, which causes the synthesis of chemokines and cytokines. These chemokines and cytokines promote neutrophil recruitment and lead to antigen-specific immunity. Dectin-1 thus plays a critical role in the start of fungal pathogen defense [18]. The Tyr238X polymorphism causes defective surface expression of dectin-1, which impairs neutrophils’ ability to destroy Candida albicans and prevents monocytes and macrophages from recognizing β-glucan and mounting a proper cytokine response [39]. Our investigation of dectin-1 rs16910526 polymorphism demonstrated significant associations with fungal coinfections in COVID-19 patients.
Notably, individuals harboring the GG genotype of dectin-1 rs16910526 exhibited a markedly elevated risk of fungal coinfections compared to those with the TT genotype. Additionally, the G allele of dectin-1 rs16910526 was strongly associated with increased susceptibility to fungal coinfections in COVID-19. Also, recessive and dominant models revealed significant associations with FI. These results underscore the importance of dectin-1 in antifungal immunity and its potential role in mediating fungal susceptibility during COVID-19. Several studies have analyzed the association between dectin-1 and its coding gene (CLEC7A) SNPs and fungal infections [40]. Plantinga et al. [41] discovered a significant incidence of aspergillosis when the SNP rs16910526 was present in both donors and patients after stem cell transplantation. Furthermore, studies found that patients receiving stem cell transplantation with the SNP rs16910526 in CLEC7A were more sensitive to Candida species colonization than those with wild-type CLEC7A (OR = 11.9, 95% CI: 2.5-56.8) [20, 42]. By contrast, Fischer et al. [21] found no link between harboring the SNP rs16910526 and pulmonary invasive fungal disease in patients with acute myelocytic leukemia (OR = 0.7, 95% CI: 0.2-2.5, p = 0.65).
The findings of the current study revealed significant associations between serum TLR2 and dectin-1 levels with genotypic variations in COVID-19 patients. Specifically, the GG genotype of dectin-1 (rs16910526) demonstrated notably higher serum dectin-1 levels compared to the TG and TT genotypes. These findings align with previous research that has suggested dectin-1’s role in modulating immune responses during viral infections, where the GG genotype may enhance cytokine production, thus contributing to severe outcomes in COVID-19 [43]. Additionally, the elevated TLR2 levels in the GG genotype (rs5743708) are consistent with studies indicating that TLR2 upregulation can enhance the innate immune response, potentially aggravating inflammatory damage in severe cases of COVID-19 [25]. However, the lack of significant differences in TLR2 levels between AA and AG genotypes suggests the complexity of host-pathogen interactions and the need for further exploration of genetic factors in COVID-19.
Despite the strength of the study, which is being one of the few investigations into the association of TLR2 (rs5743708) and dectin-1 (rs16910526) gene polymorphisms with fungal co-infection in COVID-19, it is not free from limitations. First, the sample size was not large enough. Second, it lacked a healthy control group. Third, the species of fungi were not identified through VITEC or sequencing technique. The final limitation of this study is the lack of a group of non-COVID-19 patients with systemic fungal infection, which can be considered a drawback.
Conclusions
The SNPs TLR rs5743708 and dectin-1 rs16910526 are associated with greatly increased vulnerability to fungal infection in COVID-19 patients. By elucidating the role of specific SNPs in the TLR2 (rs5743708) and dectin-1 (rs16910526) gene polymorphisms, we can gain insights into how these genetic variations impact the host immune response to fungal pathogens. This knowledge can aid in the development of personalized treatment approaches, such as targeted antifungal therapies, for individuals with these genetic variants. The levels of serum TLR2 and dectin-1 can be influenced by variations in the corresponding genotypes. For example, individuals with GG genotypes of TLR2 rs5743708 have higher levels of the corresponding protein in their serum. Similarly, variations in the genotypes of dectin-1 (GG genotype) can also affect the rising levels of dectin-1 protein in the serum. These variations in genotype can have implications for the functioning of the immune system and may contribute to differences in immune responses and susceptibility to COVID-19. Additionally, subsequent epidemiological research can determine the frequency of these mutations in different local populations and assess whether they are associated with an increased risk of fungal infections. Identifying such genetic variants related to candidiasis or aspergillosis can help identify high-risk groups and guide preventive measures or early interventions. Ultimately, this investigation has the potential to improve patient outcomes and reduce the burden of fungal infections in COVID-19 patients.