Introduction
Currently, cancer imposes a huge global health and economic burden, and many tumours have poor survival rates. Liver cancer is one of the most common malignant tumours and is difficult to detect in the early stage, with strong metastasis and poor prognosis. It is already one of the leading causes of cancer-related death, and its incidence continues to increase. Many patients are effectively diagnosed and treated at an early stage of the disease, and their 5-year survival rate can be as high as 70% [1]. Patients with advanced disease, despite treatment, have a poor prognosis, with a 5-year survival rate of only 11% [2]. In recent years, although the treatment methods for liver cancer have gradually improved, the overall survival rate is still low, especially for patients with advanced liver cancer. Due to the lack of specific symptoms and diagnostic markers for early diagnosis, liver cancer is not easy to detect in the early stage. Therefore, new diagnostic markers for liver cancer are urgently needed, to improve the current environment for liver cancer treatment [3, 4].
Anti-silencing function 1 (ASF1) was originally discovered in yeast as a histone H3-H4 chaperone. ASF1 plays an important role in the process of histone polymerization, transport, and modification. ASF1 has 2 main isoforms: ASF1A and ASF1B [5, 6]. The ASF1b protein, a substrate of the tangle-like kinase family of cell cycle-regulated protein kinases, plays a key role in regulating the nucleosomal structure of chromatin through the continuous supply of histones to the site of nucleosome assembly, while also participating in the role of transcriptional regulators [7]. Previous studies have shown that ASF1B affects the pathogenesis of cancers such as cervical cancer, prostate cancer, and clear cell renal cell carcinoma, and it can enhance tumour cell growth and migration [8–10]. Another study reported the role of ASF1B in human B-cell proliferation by promoting S-phase entry and mitotic progression [11]. However, there are still few reports on the expression and function of ASF1B in LIHC. The current study only reported that ASF1B may be a target to inhibit the growth of hepatocellular carcinoma cells [12].
Aim
This study aimed to evaluate the diagnostic and prognostic significance of ASF1B expression in human LIHC on the basis of TCGA data. The biological processes of ASF1B were explored through meta-analysis and gene enrichment analysis to assess the general prognostic significance of ASF1B. The expression of ASF1B in 33 different human cancers and its relationship with immunomodulators, immunotherapy, and dynamic immune biomarkers were also investigated.
Material and methods
Data collection
RNA-seq data of liver cancer samples and non-tumour samples were collected from the TCGA (https://cancergenome.nih.gov/) dataset. The data were ID-transformed to obtain expression matrices, and the “Limma” package of R studio software was used to convert the The expression level of ASF1B gene in the matrix was extracted. The other data included LIHC RNA-seq (GSE76427) of 115 LIHC patients from GEO (https://www.ncbi.nlm.nih.gov/geo/), and the clinical data of LIHC patients used in this paper were obtained from TCGA and GEO. Expression data, mutation data, survival data, and pan-cancer clinical data information for 33 cancers were obtained from UCSC Xena: (http://xena.ucsc.edu/). A total of 463 clinical data were obtained, and age, grade, M, N, T, gender, and stage were selected for subsequent analysis. For the treatment cohort, a systematic search was performed to identify immune checkpoint blockade cohorts that can be publicly retrieved and reported with full clinical information. These data were pre-processed using R software.
META analysis
A comprehensive search of published articles on ASF1B and prognosis of liver cancer in PubMed, Google Scholar, Web of Science, and Embase found only a few articles [12, 13]. For this reason, we collected information from the databases of TCGA and GEO to evaluate the prognostic value of ASF1B in LIHC patients. Composite HRs and 95% CIs were calculated to assess the association of ASF1B expression with prognostic outcomes in LIHC patients. R language was used for prognostic meta-analysis. If I2 was greater than 50% and p > 0.05, a fixed-effects model was used; otherwise, a random-effects model was used.
Functional and clinical and immune correlation analysis
To understand the potential role of ASF1B in LIHC patients, we performed GO and KEGG analysis using the ClusterProfiler package in R, with p less than 0.05 considered statistically significant. Using the limma package in R studio software, the expression level of the target gene ASF1B was extracted and differentially analysed to observe whether it was different in normal tissue and tumour tissue. We also investigated whether ASF1B expression differs between different clinical groups (sex, age, tumour stage and grade). Gene activity scoring was performed using the limma, GSVA, and GSEABase packages in R studio software. According to the expression of target genes, they were divided into high- and low-expression groups, and Kaplan-Meier survival analysis was performed using the survival package in R to study the time-dependent value of ASF1B in 33 cancers. The content of the study included overall survival (OS: time from the start of treatment to death), disease-free survival (DFS: the time from the start of treatment to disease recurrence), disease-specific survival (DSS: the change of the outcome measure from a specific death due to disease), and progression-free survival (PFS: the period from initiation of treatment to progression of disease). A p-value < 0.05 was considered to be a difference in survival between the high- and low-expression groups. Then the correlation analysis of immune genes was carried out. In addition, the relationship between ASF1B expression and immune suppressive genes, immune stimulation genes, and MHC-related genes was explored through the TISIDB website (http://cis.hku.hk/TISIDB/index.php). This study analysed 4 relevant independent immunotherapy cohorts to investigate whether the expression of the target gene ASF1B affects the efficacy of immunotherapy in patients. In this study, those who achieved complete remission and partial remission were classified as the responding group, and those who achieved disease progression and stable disease were classified as non-responding group, and then the 2 groups were compared.
Statistical analysis
Statistical analysis of all data in this paper was performed by R program 4.1.2. Diversity between groups was assessed by the Mann-Whitney test, and categorical data were analysed by the χ2 test. Kaplan-Meier survival analysis was performed by the survival package in R and evaluated by the log-rank test. Linear regression analysis was used to explore clinical and immunological correlations. Time-dependent ROC curves were used to determine the prognostic effect by comparing the AUC with the R package “pROC”. Analysis of variables was done by Cox proportional hazards regression analysis. P less than 0.05 was considered statistically significant.
Result
Up-regulation of ASF1B in hepatocellular carcinoma and its clinical significance
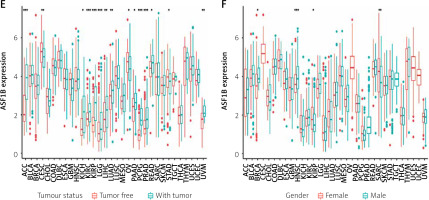
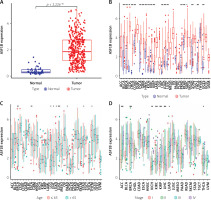
The 33 cancers involved in this study are shown in Table I. Analysis of the TCGA dataset found that the expression of ASF1B was significantly increased in LIHC samples compared with non-cancer samples (Figure 1 A), indicating that ASF1B was highly expressed in liver cancer tissues, and expressed in both liver cancer and normal tissues. have a statistically significant difference. At the same time, in Figure 2 B ASF1B is present in 33 tumours in addition to LIHC, but also in BLCA, BRCA, CESC, CHOL, ESCA, GBM, HNSC, KIRC, KIRP, LUAD, LUSC, PRAD, READ, SARC, STAD, THCA , and UCEC, the expression was different in these tumours. In Figures 1 C–E, in the clinical correlation analysis, dysregulation of ASF1B expression was associated with LIHC patients in age, tumour stage, and tumour and normal tissues, regardless of gender (Figure 1 F).
Table I
The 33 cancers used in this study
Figure 1
Significant upregulation of ASF1B and its association with clinical features. A – ASF1B expression was determined in LIHC specimens and non-tumour specimens using the TCGA dataset. B – The differential expression analysis of the ASF1B tumour group and normal group in 33 cancers. C – The correlation between age and ASF1B. D – The correlation between tumour stage and ASF1B. *p < 0.05, **p < 0.01, ***p < 0.001
Correlation analysis of ASF1B expression and prognosis of LIHC patients
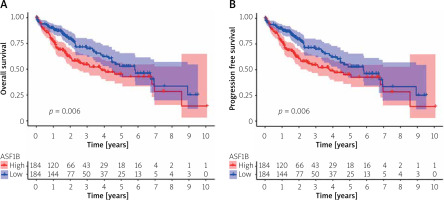
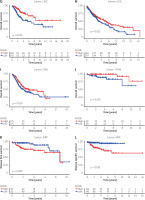
To explore the prognostic value of ASF1B in liver cancer, first, according to the results, the overall survival (Figure 2 A), progression-free survival (Figure 2 B), disease-specific survival (Figure 2 C), and disease-free survival (Figure 2 D) were studied. Patients with high expression levels of ASF1B had a low overall survival rate, indicating that patients with high ASF1B had poorer survival. Subsequent analysis of related ROC curves showed that the AUCs of 1-year, 3-year, and 5-year survival were 0.672, 0.590, and 0.591, respectively (Figure 2 E), and the values were all greater than 0.5, indicating that the prognostic indicator based on ASF1B gene expression had a significant effect on survival. Next, Figure 2 F shows that the univariate independent prognostic analysis reveals that ASF1B is the only factor associated with the prognosis of LIHC patients among the many factors (HR = 1.288, 95% CI: 1.085–1.529, p < 0.004), further indicating that ASF1B plays a role in the prognosis of HCC patients. Finally, in Figure 3, we also analysed the effect of ASF1B on overall survival in 33 human cancers, and found that in addition to the LIHC patients studied in this paper, in ACC, KIRC, LGG, LUAD, MESO, and PAAD the expression of ASF1B was significantly correlated with overall survival. There was a clear negative correlation with survival, but a positive correlation with CESC, LUSC, STAD, and THYM. The expression of ASF1B was negatively correlated with DFS, DSS, and PFS of KIRP but positively correlated with STAD. The prognostic role of ASF1B gene in other tumours is different, which may imply that the gene has a different mode of action or function and pathway in different tumours, which may require more research to verify.
Meta-analysis and predictive performance of ASF1B expression
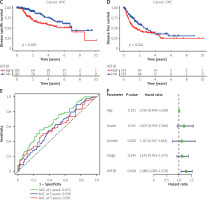
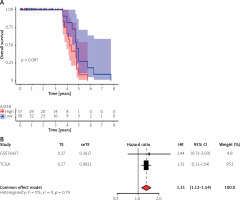
The survival significance of ASF1B expression was analysed using GSE76427. Unfortunately, no clear association of dysregulated ASF1B expression with LIHC patients was observed, but patients with high ASF1B expression showed a clinical trend of poorer overall survival (Figure 4 A). Next, we performed a meta-analysis using 115 LIHC patients from GSE76427 and the TGCA dataset. As shown in Figure 4B, the pooled HR (1.32) and 95% CI (1.12–1.54) of the relationship between ASF1B expression and overall survival showed no significant heterogeneity between the 2 datasets. Therefore, we can conclude that patients with high ASF1B expression can improve the effective prediction of survival of HCC patients. The higher the expression of ASF1B, the worse the prognosis of liver cancer patients.
Analysis of ASF1B-related functional and immune correlates in LIHC
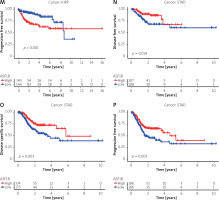
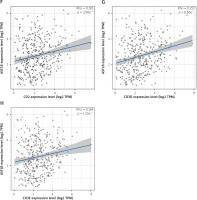
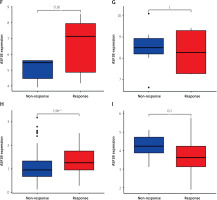
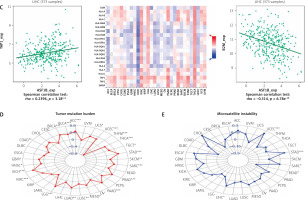
The dysregulated genes in the LIHC samples of the ASF1B high-expression group were first screened, and then GO and KEGG analyses were performed using these genes. In Figure 5 A, GO enrichment analysis indicated that the biological processes of differential genes were mainly involved in chromosome segregation, nuclear division, organelle fission, and chromosomal region. KEGG enrichment analysis (Figure 5 B) showed that the main pathways of differential genes were cell cycle, retinol metabolism, metabolism of xenobiotics by cytochrome P450, and drug metabolism-cytochrome P450. As shown in Figure 5, we also analysed the correlation of ASF1B with immune cells (C–E) and immune genes (F–H), and found that it was positively correlated with B cells, CD8+ T cells, and regulatory T cells, and also with CD2, CD3D and CD3E Isoimmune genes were positively correlated. The correlation between ASF1B expression and immunomodulators was also investigated. In the analysis of immunosuppressants, Figure 6 A shows that the expression of ASF1B is positively correlated with LAG3 of LIHC and negatively correlated with KDR. Figure 6 B shows that in the analysis of immunostimulants, ASF1B expression was positively correlated with MICB of LIHC and negatively correlated with CXCL12 of LIHC. It shows that ASF1B has a certain role in immunity and is affected by immune stimulation or immunosuppression, which can provide certain new ideas and directions for the immunotherapy of liver cancer patients. In Figure 6 C, in the analysis of MHC-related genes, it was found that ASF1B expression was positively correlated with TAP1 of LIHC, but negatively correlated with B2M of LIHC. In Figure 6 D, we also performed a correlation analysis of tumour mutational burden, and the results showed that the expression of ASF1B was associated with ACC, BLCA, BRCA, GBM, KICH, KIRC, LGG, LUAD, PAAD, PRAD, SARC, SKCM, STAD, and THCA. The mutational burden of these tumours was strongly correlated with UCEC and THYM, but unfortunately no correlation with LIHC was found. Next, we performed a correlation analysis of microsatellite instability (Figure 6 E), and the expression of ASF1B was positively correlated with BLCA, ESCA, LIHC, SARC, STAD, and UCEC but negatively correlated with READ. Finally, we performed a correlation analysis of ASF1B immunotherapy response (Figures 6 F–I), in the IMvigor210 immunotherapy cohort; we found that ASF1B was highly expressed in the responder group, while in the 3 immunotherapy cohorts (GSE157893, GSE67501, and GSE78220), there was no significant difference in the expression of FOXP3 between the responder group and the non-responder group. The results show that in pancancer, ASF1B's response to immunotherapy is only statistically different. Provide new directions for chemotherapy.
Figure 5
A – Results of GO enrichment analysis. B – Results of KEGG enrichment analysis. C–E – The relationship between ASF1B and immune cells B cells, CD8+ T cells, and regulatory T cells

Figure 6
A – Correlation of ASF1B expression with immunosuppressive agents. B – Correlation of FOXP3 expression with immunostimulatory agents
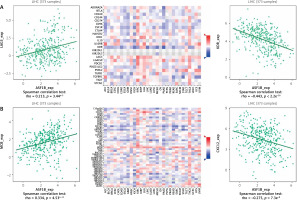
Figure 6
Cont. C – The expression correlation of ASF1B and MHC molecules. Red indicates positive correlation; blue indicates negative correlation. D – Correlation of ASF1B with immunotherapy markers in tumour mutational burden. E – Aspects of microsatellite instability correlation analysis. *p < 0.05, **p < 0.01, and ***p < 0.001
Discussion
Because of its insidious onset and rapid progress, liver cancer is mostly in the advanced stage when diagnosed. Therefore, early diagnosis and early treatment have become important parts of the clinical treatment of liver cancer today [14, 15]. Early clinical screening methods were mainly serum α-fetoprotein (AFP) and liver imaging. However, the sensitivity and specificity of AFP detection are limited, and some physiological activities can affect the changes of its indicators, and imaging examinations are difficult to effectively identify early space-occupying lesions in the liver. Therefore, screening for molecular biomarkers that can fully reflect the biological characteristics of liver cancer is a key link in the early diagnosis and intervention of liver cancer patients. The establishment of a multi-molecular biological prediction model of liver cancer has very important clinical significance for improving the treatment effect and prognosis [4, 16]. The ASF1B gene is located on chromosome 19q13.12 and consists of 4 exons that regulate the nucleosome structure of chromatin by encoding members of the histone chaperone H3/H4 family, and providing histones to nucleosome assembly sites plays a key role in [8, 17, 18]. It has been reported that ASF1A and ASF1B in humans can co-deplete the alternately elongated telomere (ALT) pathway maintained at the ends of mammalian chromosomes in primary cells and cancer cells [19]. ASF1B is an important oncogenic regulator in the development and progression of lung adenocarcinoma and other types of cancer [20], and it has also been shown to play a key role in local and metastatic breast cancer recurrence and patient prognosis [21, 22].
Here, our results based on the TCGA dataset showed that ASF1B was significantly upregulated in LIHC compared with non-tumour patients. Pan-cancer analysis also showed that in addition to LIHC, ASF1B was present in 33 tumours (BLCA, BRCA, CESC, CHOL, ESCA, GBM, HNSC, KIRC, KIRP, LUAD, LUSC, PRAD, READ, SARC, STAD, THCA, and UCEC) and was differentially expressed in these tumours. We divided all the LIHC patients into 2 groups according to the average expression of ASF1B. Survival analysis showed that patients with high ASF1B expression levels had a lower overall survival rate, including lower survival rates in progression-free survival, disease-specific survival, and disease-free survival. To further understand the impact of ASF1B on the survival of tumour patients, we also analysed the impact of ASF1B on the overall survival of 33 human cancers and found that in addition to the LIHC patients studied in this paper, in ACC, KIRC, LGG, LUAD, MESO, and PAAD, ASF1B expression was significantly negatively correlated with overall survival, while it was positively correlated in CESC, LUSC, STAD, and THYM. The expression of ASF1B was negatively correlated with DFS, DSS, and PFS of KIRP but positively correlated with STAD.
A meta-analysis involving 115 LIHC patients from GSE76427 and the TGCA dataset showed that patients with high ASF1B expression could improve the effective prediction of survival in HCC patients. In addition, GO enrichment analysis indicated that the biological processes of differential genes were mainly involved in chromosome segregation, nuclear division, organelle fission, and chromosomal region. In previous studies, Chan et al. [23] found that DEAD-box helicase 3, X-linked (DDX3X) is associated with liver injury and liver tumour development through chromosome segregation. Regarding organelle fission, high expression of the PPP2CA gene in HCC tissue is significantly associated with poor prognosis of liver cancer patients, and the PPP2CA gene and related genes in the study are also enriched in organelle fission [24]. KEGG enrichment analysis showed that the main pathways of differential genes are cell cycle, retinol metabolism, metabolism of xenobiotics by cytochrome P450, and drug metabolism−cytochrome P450. These findings may provide more ideas for future research. Regulation of the cell cycle is crucial for the growth of malignant tumour cells, and this process represents the target of anticancer therapy [25]. Pu et al. [26] found that mir-135a can block tumour cells in the G1 phase and affect the cell cycle progression, thereby affecting cell proliferation and development. In previous research, Pettinelli et al. [27] found that altered retinol metabolism is one of the pathways involved in the process of liver fibrosis. At the same time, the enzymes involved in retinol metabolism are related to the occurrence and development of liver cancer.
In order to further study the potential value of ASF1B, the correlation between ASF1B and immune cell infiltration was explored, and it was found to be positively correlated with B cells, CD8+ T cells, and regulatory T cells, as well as with immune genes such as CD2, CD3D, and CD3E. Hirsova and Gores [28] found that CD8 T cells play a key role in causing liver damage and promoting the development of hepatocellular carcinoma in nonalcoholic steatohepatitis. Regarding regulatory T cells, previous studies have confirmed that regulatory T (Treg) cells play a crucial role in maintaining an immunosuppressive tumour microenvironment. TGF-β signalling in sexual T cells to promote tumourigenesis and development. The expression of ASF1B was positively correlated with LAG3 of LIHC in the analysis of immunosuppressant and negatively correlated with KDR. In the analysis of immunostimulants, ASF1B expression was positively correlated with MICB in LIHC but negatively correlated with CXCL2 in LIHC. The expression of ASF1B was positively correlated with TAP1 of LIHC in the analysis of MHC-related genes and negatively correlated with B2M. In terms of the correlation analysis of tumour mutation burden and microsatellite instability, we performed a pan-cancer analysis of 33 human cancers. Unfortunately, in terms of tumour mutation burden, ASF1B was observed to be associated with LIHC, while in ACC, BLCA, BRCA, GBM, KICH, KIRC, LGG, LUAD, PAAD, PRAD, SARC, SKCM, STAD, THCA, UCEC, and THYM tumours were strongly associated with mutational burden, but in microsatellite instability analysis the expression of ASF1B was positively correlated with BLCA, ESCA, LIHC, SARC, STAD, and UCEC but negatively correlated with READ. In the correlation analysis of immunotherapy response, we found that in the IMvigor210 immunotherapy cohort ASF1B was highly expressed in the response group, which may provide new ideas and methods for the immunotherapy of liver cancer. Our findings underscore the potential of ASF1B as a biomarker in HCC patients. At the same time, this paper has certain limitations. Because only the TCGA database has PFS data, it is difficult to complete the meta-analysis of PFS. Although preliminary studies on the biological process of ASF1B in LIHC have been completed by enrichment analysis, more experiments are still needed [29].
Conclusions
Our findings show that ASF1B is significantly elevated in LIHC, and both the development of cancer and poor prognosis are associated with high expression of ASF1B, so it may be an oncogene in the pathogenesis and progression of liver cancer. We also revealed the immune value of ASF1B through a pan-cancer analysis of 33 human cancers, which suggests that ASF1B may not only be a prognostic biomarker but also a potential therapeutic target for liver cancer. These findings will provide new ideas and directions for the follow-up study of liver cancer.