Introduction
In the last decade, many microorganisms have been progressively linked in the pathogenesis of inflammatory bowel disease (IBD) – mainly Crohn’s disease (CD). Escherichia coli and particularly the adherent invasive E. coli (AIEC) pathotype, has been increasingly implicated in the etiopathogenesis of CD, but until today the mechanism of AIEC in the pathogenesis of CD remains uncharted. Significant progress has been made in recent studies on explaining the pathogenicity mechanisms of AIEC in CD patients. At the molecular level, however, the characteristics of this strain remain controversial.
The human gut microbiota composes a population of approximately 1014 commensal microorganisms. Their genomes (also called metagenome or microbiome) are about 150 times the size of the human genome in terms of the number of genes [1]. In fact, the microbiota has an effect on the physiology and metabolism within the body. Moreover, to affecting the metabolism of the host, microbiota could also be involved in several pathological mechanisms. Activation and the development of the mucosal immune system in the gastrointestinal (GI) tract depend on the complex association of these microorganisms [2]. Recent studies have linked to the role of the human gut microbiota in several human diseases, such as colon cancer, IBD, type 1 diabetes, insulin resistance, non-alcoholic fatty-liver disease, allergies, and asthma [3]. Therefore, it is very important to understand the connection of the microbiota in the aetiology of such diseases by distinguishing species that compose a healthy microbiota [4–9].
The influence of the microbiota on human health is best demonstrated by studies in IBD, such as CD and ulcerative colitis (UC) [10–12]. Both CD and UC represent serious medical disorders noticeable by abnormal inflammation within the human GI tract, which results in serious clinical outcomes in affected patients. These diseases are very complex and involve the contribution of genetic factors as well as external factors such as geographical area [13]. IBD is caused by a dysfunction of the human immune response to gut microbiota and occurs in the case of host genetic susceptibility. CD is a chronic and commonly incapacitating inflammatory intestinal disorder, whose prevalence and incidence are on the rise in developed countries [14].
The aim of this review is to discuss recent findings that indicate the role and mechanisms of AIEC in IBD, and to highlight the AIEC virulence factor genes and their mechanisms that suggest playing a crucial role in the severity of inflammation in CD patients. It is also aimed to discuss the current data on the prevalence of AIEC in CD patients.
The role of Escherichia coli in inflammatory bowel disease
Escherichia coli strains have been classified into seven (A, B1, B2, C, D, E, and F) phylogenetic groups according to virulence factors. Recently, the pathogenesis of IBD has been linked to human intestinal microbiota [15]. Patients with CD display an altered gut microbial community, and the imbalance (dysbiosis) present in patients with colonic and ileac CD is different [16]. In contrast, a specific gut microbiota imbalance is beginning to be identified in UC patients, but differences between studies have inhibited attempts to reach a clear conclusion to date [16–19]. E. coli is the most prominent bacterium in CD aetiology in the last 10–15 years [20, 21] (Table I) [22–30]. The growth of the E. coli population in IBD patients is currently unexplained, but that may be due to the association with increased production of reactive nitrogen species, allowing nitrate respiration, which confers E. coli a fitness advantage [31]. Recent studies based on cultural and molecular techniques support the theory that E. coli is an important microbial factor involved in CD pathogenesis, but some disagreement exists regarding its role in UC pathogenesis [17–19, 32–34]. In this section, we examine the recently published data on E. coli populations in CD patients related to their abundance, consortium associated with disease activity, and alteration of the human gut mucosa. We have also focused on the pathogenic properties of the strains to highlight evidence supporting or limiting the inclusion of this bacterium into the IBD subtype.
Table I
Abnormal prevalence of E. coli in Crohn’s disease patients in the last 5 years
Recently, an elevated number of mucous AIEC strains have been isolated from the gut mucosa of CD patients [35–38]. This has led to the illustration and identification of a new bacterial strain known as AIEC, which is characterised by its particular capacity to adhere and invade the cells of human small intestines, especially ileal cells [33, 39].
The most common virulence factors in adherent-invasive Escherichia coli strains
As mentioned previously, AIEC strains were extensively linked to many aspects of CD pathogenesis, and their virulence factors were compared in the reference AIEC LF82 strain and non-AIEC strains. Despite all the research on AIEC pathogenicity, the exact genetic factors that could define it as a feature of the AIEC prototype are still unknown. The majority of genes studied for AIEC pathogenicity are not AIEC-specific genes as for the fimH, htrA, dsbA or ompA genes, and these genes are also found in most E. coli strains, including non-pathogenic E coli [40–43].
E. coli possess fimbriae as a virulence factor, which confers pathogenic strains with the ability to adhere to and colonise various specific host epithelia. So far, the fimH gene is one of the most studied virulence factors in AIEC strains. The FimH gene encodes an adhesion characteristic that allows the bacterial adhesion to glycosylated and non-glycosylated host receptors, as well as the matrix-associated type I and IV collagens, glycosylated receptors, fibronectin, and laminin [44]. Even though almost all E. coli strains comprise type 1 pili including non-pathogenic strains, most AIEC strains generally offer a variation of fimH adhesion, which makes them more effective for binding to human intestinal epithelial cells [40]. Some other non-AIEC strains possess these mutations as well, but these strains do not exhibit type 1 pili. Mutation of the fimH gene in AIEC strains can increase the ability to adhere to expressed carcinoembryonic antigen-related cell adhesion molecules (CEACAM6) in intestinal epithelial cells (Figure 1). The expression of type 1 pili and long polar fimbriae (LPF) of AIEC strains allows target M cells on Peyer’s patches, which help to translocate across the barrier of intestinal epithelial cells [3]. LPF is considered a pathogenic feature and is one of the characteristics of AIEC strains in CD (Table II) [45–61].
Figure 1
The diagram shows changes in the gut during inflammation mediated by adherent-invasive Escherichia coli (AIEC) colonisation. Inflammation of the human gut can be triggered by several factors including diet, antibiotic administration, acute gastroenteritis, and host genetics. The pro-inflammation in human gut mediates noticeable changes in the gut, which can appear as hypoferremia and over-expression of carcinoembryonic antigen-related cell adhesion molecule (CEACAM6) surface receptors produced by epithelial cells in human gut. AIEC strains have been evolved and gained a competitive advantage in inflamed gut of the human intestines. This evolution includes the ability of AIEC strains to use a modified f imH protein to bind to CEACAM6 and to be able to utilise amine (N) and sulphur (S) oxides as electron acceptors
AIEC – adherent-invasive Escherichia coli, CEACAM6 – carcinoembryonic antigen-related cell adhesion molecule, Th – T helper, IL – interleukin, TNF-α – tumour necrosis factor α, IFN-γ – interferon γ.
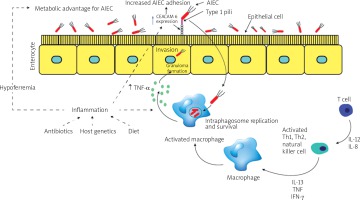
Table II
Virulence factors described in AIEC strains
Nevertheless, recent studies have shown that the fimH gene does not show any significant association with AIEC pathotype; this gene has been detected in both AIEC and non-AIEC strains [30]. However, a higher rate of the fimH gene has been reported in AIEC strains isolated from biopsy samples of patients with UC compared to control subjects [62]. The FimH gene is the only gene that has been detected in all AIEC strains, mainly in B2 phylogenetic group E. coli [29]. The fimH gene and its protein were found to play an important role in the binding of E. coli to human epithelial colorectal adenocarcinoma (Caco-2) cells. A higher similarity between fimH alignments in the B2 phylogroup and adhesive strain of E. coli has been detected [63].
Invasion of the brain endothelium protein A (ibeA) gene; some extra-intestinal pathogenic E. coli (ExPEC) strains from phylogenetic group (B2) and especially new-born meningitis and avian pathogenic strains carry this virulence gene. The ibeA gene has invasion characteristics that allow E. coli K1 to invade blood brain barrier. The ibeA gene was originally characterised and cloned from the chromosome of an invasive E. coli K1, which has been isolated from cerebrospinal fluid [64]. E. coli K1 carrying the ibeA gene is thought to contribute to the pathology of neonatal meningitis E. coli (NMEC) and is responsible for most cases of meningitis in neonates [64, 65]. NMEC strains harbour the ibeA gene, which is associated with reductive evolution, indicating a high degree of protection [66]. This gene is responsible for the interaction of AIEC strains with human intestinal epithelial cells and macrophages, as well as colonisation of the mouse intestines.
Several studies have been conducted to identify virulence factors associated with the AIEC phenotype. The ibeA virulence gene, which plays a role in invasion, was more common in the E. coli isolates in CD patients than in controls [67]. However, ibeA gene has been also detected in the genome of AIEC strain (NRG857c), and this gene contributes to the invasion, macrophage survival, and inflammatory response in the murine intestine [68].
Regarding to the molecular mechanisms mediating ibeA gene interactions with host cells, existing data are quite insufficient. However, this gene was reported as a 50-kDa outer membrane protein containing seven predicted trans-membrane domains with expanded layers passing from the cell membrane to the extracellular space [68]. According to recent reports, the ibeA gene can bind host proteins as potential receptors, including polypyrimidine tract-binding protein (PTB)-associated splicing factor (PSF), a RNA-binding component of spliceosomes, and vimentin (a type III intermediate filament (IF) protein that is expressed in mesenchymal cells) [27, 69].
The ibeA gene encodes an RNA polymerase and sigma S (RpoS)-like regulator with a narrow functional spectrum, which is considered to play a part in bacterial virulence adaptation in some NMEC strains, and this gene also located in the same operon with other genes (ibeR and ibeT) [70]. It is not known whether i beA is regulated by ibeR, but brain endothelial cell invasion is affected by the absence of ibeA [70]. However, the ibeT gene is located near to ibeA and has been linked to affect invasion and adhesion of brain endothelial cells, even though it shows sequence homology to Na+/H+ antiporters [71].
Polysaccharide K capsule gene – many pathogenic E. coli strains, including AIEC strains, carry a polysaccharide K capsule that protects the bacterium against host innate immune factors, and it plays a major role in resistance and survival during infection. The pathogenic ExPEC strains express a polysaccharide capsule that is important both pathogenically and taxonomically [72, 73]. Commensal E. coli carries a high-molecular-weight, low-charge-density group 1 capsule; in contrast, ExPEC carries low-molecular-weight, high-charge-density group 2 and 3 capsules that protect ExPECs against phagocytosis and complement-mediated killing, thereby contributing to extra-intestinal virulence [72, 74–78]. Nevertheless, the characterisation of this K antigen is highly specialised and cannot be performed in any laboratory. In contrast, molecular detection of kps antigens can be performed in any molecular laboratory. Kps 2 and 3 operons share moderate and highly conserved regions that encode transport and assembly functions, which combine with type-specific regions that encode the synthesis of the specific component sugars of the particular polysaccharide [79–84]. The contribution of capsular antigen is well established in uropathogenic E. coli (UPEC) and protects against phagocytosis and complement-mediated killing [85, 86].
In laboratory animals such as mice, K1 contributes to the development of intracellular bacterial communities that are biofilm-like bacterial aggregates in superficial bladder epithelial cells during the early stages of acute urinary tract infections (UTI) [87]. K1 serotype is highly associated with bacterial strains that cause blood infection, meningitis, and UTI [86, 87]. K1 capsule is made from sialic acid chain residues that are synthesised by enzymes encoded by genes in region II of the capsule locus (neuDBACES), and this polysaccharide is similar to the polysialic acid found on some human cells, and due to molecular mimicry the K1 antigen is considered poorly immunogenic [88].
K1, K5, and kpsMT II genes are involved in the synthesis of capsular materials and are important in the virulence of bacteria. These genes have recently been identified in the AIEC LF82 strain [89–92]. A positive association between capsular genes (K1 and kpsMT II) in paediatric patients with CD has also been detected [67]. The importance of these genes and their presence only in CD patients could additionally support that AIEC strains may persist in CD.
Virulence gene ferric hydroxamate uptake protein D (fhuD), outer membrane hemin receptor (chuA), and iron-regulatory proteins (irp2) are suggested to be related with iron uptake. Iron is an essential element for all microorganisms, except some Lactobacilli. In fact, total iron in the human body amounts to about 4–5 g and it is not readily available for the bacteria because the iron is bound to eukaryotic proteins such as ferritin, transferring, haemoglobin, and lactoferrin. A host with a poor iron environment is a clear sign that influences the mechanism of iron acquisition. A low-molecular-weight Fe3+ binding compound (siderophores) will be synthesised by the bacteria to transport and solubilise the iron to the bacteria, and Fe3+-siderophore complex is brought into the cell by a membrane protein [93]. Pathogenic bacteria have developed different mechanisms to obtain and compete with the host for iron, which is an essential growth factor for the host and the bacteria [94]. However, a lack of iron generates the expression of some virulence factors such as toxins [95–97] and haemolysins [98–100].
The fhuD gene; E. coli possesses a ferric hydroxamate transport system, which is a soluble periplasmic binding protein. Fe3+ cannot be transported a mono-atomic ion because of its extreme insolubility. Iron is bound to low-molecular-weight carriers designated siderophores in microbes. E. coli Fe3+ siderophores have to be translocated across two membranes for uptake into the cells. After conversion of E. coli cells to spheroplasts fhuD protein is released, which indicates a location in the periplasmic space between the outer membrane and the cytoplasmic membrane [101]. The properties of this protein are typical for bacterial periplasmic binding protein-dependent transport systems (PBPs) through which peptides, amino acids, anions, vitamins, and some sugars are absorbed [101].
The chuA gene is a haeme iron acquisition gene. Several pathogenic E. coli strains carry this gene as an outer membrane protein responsible for haemin utilisation. A Recent study identified the chuA gene, which encodes the 69-kDa outer membrane protein responsible for haeme uptake in E. coli O157:H7 [102]. This gene is part of the haeme transport locus, which is widely distributed among pathogenic E. coli strains [103]. Iron is an essential element for growth of AIEC, and the AIEC strains that are enriched with siderophores (chu operon) are able to survive and persist inside J774 macrophages, which can be suggested as a major contributor to acquiring and using iron and could encourage the multiplication of AIEC in inflamed human intestines [53]. Within macrophages, AIEC could selectively utilise the host defects in autophagy [104], by upregulating of the chuA gene and stimulating the release of TNF-α to promote and enhance dysbiosis, multiplication, and persistence of AIEC [105, 106].
The irp2 gene regulates the post-transcriptional expression of mRNAs that encode certain proteins involved in iron utilisation and homeostasis [107–109]. Disorders of iron metabolism can cause major health problems because iron is a very important element for cellular functions. It can be linked to some metabolic processes, such as cell growth, inflammation, and apoptosis. Vertebrate iron metabolism and transcriptionally regulated expression of the major iron homeostasis genes are controlled by iron-regulatory proteins-2 (IRP2) [110]. Recent studies have shown that virulence genes related to iron uptake (fhuD,chuA, and irp2) have been detected in AIEC strains in patients with CD [30, 53]. Pathogenic and non-pathogenic E. coli strains carry the fhuD gene; however, the chuA and irp2 genes are less common among diarrhoeagenic and commensal E. coli strains [30]. Nonetheless, ExPEC strains carry both these genes. The presence of fhuD, chuA, and irp2 genes or the presence of just the chuA gene alone may play a crucial role in detecting AIEC in patients with CD [30]. This can be a suitable biological and diagnostic marker that can be used to identify and characterise AIEC strains in CD patients.
Ferric yersiniabactin uptake receptor (fyuA) gene acts as a receptor for iron yersiniabactin (Fe-Ybt) siderophore uptake [111–113]. The virulence in many members of the Enterobacteriaceae family has been linked to this gene [113, 114]. This gene not only acts as a siderophore uptake receptor but is also involved in biofilm formation. Nevertheless, the role of the fyuA gene in biofilm formation is not known yet, nor whether the effect is due to a decrease in concentration of intracellular iron or as a result of another mechanism that remains to be explained [115]. The fyuA gene has been identified in AIEC-like strains isolated from CD patients [67]. Pathogenic bacteria grow and multiply by using Hb or haem as the sole iron source. These pathogenic bacteria obtain an entrance to the intracellular haeme reservoir alongside starting tissue invasion by secreting certain cytotoxins. During the progression of infection in CD patients, cytotoxin production combined with the haeme or/and haemoglobin using capacity to could assist the effect of iron acquisition.
The high-pathogenicity island (HPI) present in pathogenic Yersinia, which carries genes like fyuA that are involved in the transport, regulation, and synthesis of the siderophore yersiniabactin, has been detected in various strains of E. coli [116]. However, HPI has been detected in verotoxin-producing E. coli and does not appear to contribute to pathogenicity, but it can contribute to the strain fitness of E. coli [117]. Furthermore, E. coli strains carrying HPI have been linked to diarrhoea in humans [118].
Ribosome-association toxin (ratA) gene is encoded by the E. coli genome [119]. This toxic protein inhibits the initiation of translation by associating specifically with the ribosome (50S subunit) and also inhibiting 70S ribosome formation. This gene has no effect on cellular mRNAs and is unable to dissociate 70S ribosomes [58]. Inducing expression of the ratA gene causes inhibition of cell growth [58, 119]. A recent study has shown a positive association between rat gene and patients with CD, and the detection of this toxic gene is likely to play a crucial role in AIEC pathogenesis in patients with CD [30]. This gene was isolated and identified from adherent and invasive strains isolated from the ileum of patients with Crohn’s disease [120].
Quantitative real-time PCR-based analysis for adherent-invasive E. coli
AIEC strains are genetically variable, and the virulence factors are nonspecific. Recent studies have linked the AIEC strains with CD [121–124]. At present, time-consuming techniques such as in vitro infection of cell cultures that has been used to determine the ability of AIEC strain to adhere and invade epithelial cells as well as to survive and replicate within macrophage cells are required for the assessment of the pathogenicity of AIEC strains. However, these time-consuming techniques do not enable precise quantification of AIEC strains from human samples. A fast, sensitive, and successful quantitative real-time PCR (qRt-PCR) technique is applied nowadays for identification and quantification of microbes from clinical samples [33, 37, 125, 126].
A qRT-PCR assay for quantification of the LF82 strain and total E. coli in human intestinal samples from CD patients has been reported. Targeted bacteria have been quantified, and a standard curve has been made. Proper primers were designed to ensure high specificity detection. This assay showed high specificity and robustness for the detection of LF82 strain in human intestinal tissues. Combining this technique with other techniques such as phenotypic assays (adhesion and replication in cell lines) will help in the isolation and characterisation of LF82 strain [39].
Conclusions
Considerable evidence indicates that E. coli and particularly AIEC strains are involved in the pathogenesis of CD. Although the prevalence of AIEC in the mucosa of CD patients has been reported in many studies, the abundance of AIEC varies significantly between studies. The virulence genes that relate to adhesion, invasion, capsule formation, iron acquisition, and toxin production among E. coli isolates from CD patients are thought to be major contributors for colonisation of E. coli in the GI tract. These virulence factors can define the pathophysiology of CD like intestinal inflammation, bacterial translocation through mucosa, and formation of granuloma. A decade ago, the AIEC pathotype was discovered, and ever since, studies have reported the ability of AIEC to adhere and to invade intestinal epithelial cells, as well as to persist and survive inside macrophage cells. There are also several studies focusing on the detection of AIEC mechanisms in CD pathogenicity, and epidemiological studies have been conducted on this disease, but further research is needed to confirm the role of AIEC on CD. Using time-consuming techniques to identify the AIEC pathotype is an important limitation, and molecular tools are needed. Moreover, molecular-based studies are needed to assist in the identification of the genetic elements among AIEC pathotypes, which can be a major contributor to understanding the pathogenicity of AIECs and their interaction with the host, and also could help in the detection of therapeutic agents for CD. Finally, to reach a definitive conclusion about the role of microbes and specifically the AIEC pathotype in CD development, it is necessary to identify AIEC genes related to disease pathogenesis.










