Introduction
Indonesia is one of ten countries with the largest number diabetics in Asia. Based on data from the Central Statistics Agency (BPS), Indonesia is ranked 7th among 10 countries with the highest number of sufferers, which amounts to 10.7 million people [1]. There was a significant increase in the number of people with diabetes from 6.9% to 8.5% between 2013 and 2018. Diabetes was ranked as the 3rd most common disease, with a prevalence of 17.49%. Cases of type 2 diabetes are the most common in Indonesia, which is caused by an unhealthy lifestyle [2, 3]. Patients with an uncontrolled lifestyle are at risk for other diseases as complications, such as heart attack, stroke, blindness, kidney failure, paralysis, and death. One of the most common complications of diabetes is diabetic ulcers [3].
Diabetic ulcers are infections that occur in the limbs of diabetic patients caused by the destruction of the deepest skin tissue, and in severe conditions it will require amputation [4, 5]. The spread of wounds in diabetic ulcers is caused by the large number of bacteria in diabetic ulcers. In previous research, it was found that 4 of 9 bacterium isolates from diabetic ulcer patients in Jember were identified as Alcaligenes faecalis [6]. The presence of Alcaligenes faecalis bacteria in diabetic ulcers was also reported by Huang et al. [7].
Alcaligenes faecalis is one of the bacteria in diabetic ulcers that is resistant to several antibiotics. It was reported that Alcaligenes faecalis is resistant to the antibiotics ceftriaxone, ceftazidime, cefoperazone, clindamycin, metronidazole, and ciprofloxacin [6]. Since 2019, Alcaligenes faecalis is known to have extensive drug-resistant (XDR) properties to the antibiotics given, specifically gentamicin, amikacin, ceftazidime, cefepime, ampicillin sulbactam, piperacillin/tazobactam, ciprofloxacin, imipenem, meropenem, and tigecycline [7]. The resistant nature of bacteria causes a slow infection healing process, so new antibiotics are needed to treat existing infections [4]. However, the process of producing new antibiotics is a lengthy process. According to research conducted by Agistia et al. on the effectiveness of antibiotics in diabetic ulcer patients, some bacteria that cause diabetic ulcers are resistant to the type of antibiotics used, specifically ceftriaxone, metronidazole, ciprofloxacin, ceftazidime, ampicillin-sulbactam, meropenem, ampicillin, and netilmicin [8]. Based on these findings, another alternative solution is needed that can stop the bacteria in diabetic ulcers, i.e. by using natural enemies of bacteria, commonly known as bacteriophages [9].
Bacteriophages are viruses that infect bacteria; they are the most abundant, omnipresent, and diversified biological group inhabiting Earth. They are detected in many places, such as soil, water, and in the human body (in faeces, saliva, sputum, blood, and urine) [10]. Bacteriophages have the ability to specifically infect bacteria. The virulent nature of bacteriophages against infected bacteria can be a solution to overcome the antibiotic resistance of bacteria [11–15]. Bacteriophages attach to specific receptors on the bacterial cell wall followed by injecting genetic material into the host cell and replicating in the host’s body, causing the host cell to lyse. Bacteriophages are transitional organisms that need a host, i.e. bacteria, to reproduce, so bacteriophages can to evolve their components and mechanisms in infecting bacteria [11, 14]. Based on this, therapy using bacteriophages is an alternative solution to be able to overcome the extensive drug resistance that is possessed by bacteria that cause diabetic ulcers.
Material and methods
Bacterial culture
Alcaligenes faecalis isolate from a previous study [6] was used in this study. Bacterial cells were cultured in Nutrient Agar (Merck) medium containing Peptone 5.0, meat extract 3.0, and agar-agar 12.0 (g/l) incubated at 37°C for 24 hours. Then isolate of Alcaligenes faecalis was stored in glycerol 80% consisting of 20% aquadest steril and 80% pure glycerol (Merck).
Isolation and purification bacteriophage
Bacteriophages were isolated from diabetic ulcer swab samples from patients who met the following inclusion criteria: grade I ulcer patients who had superficial and localized ulcers, and grade II ulcer patients who had deep ulcers with cellulitis without abscess/bone deformity. The swab isolate sample suspension was cultured on Luria Bertani (Merck) medium containing tryptone 10.0, yeast extract 5.0, and sodium chloride 10.0 (g/l) that had been inoculated with Alcaligenes faecalis at 37°C for 24 hours. The incubation results were centrifuged (High Speed Centrifuge Lab Mini) at 12,000 rpm for 10 minutes. The supernatant was passed through a 0.20 µm membrane filter. The results of the filtration were carried out by spot tests on top agar medium with Alcaligenes faecalis used as the primary host. Positive results are indicated by the appearance of plaque on the medium. Bacteriophages that showed positive results in the spot test were then dissolved in SM buffer (50 mM Tris-HCl at pH 7.5, 100 mM NaCl, 10 mM MgSO4, and 0.01% gelatine). Purified phages were stored at 4oC until used for the plaque assay method [15].
Host Range
A host range test was carried out using Staphylococcus aureus, Klebsiella pneumoniae, and Escherichia coli obtained from our laboratory collections with the spot test method following Narulita et al. [15]. The incubated bacterial cultures were then dissolved in 5 ml of top agar (0.25% agar) and poured into nutrient agar (Merck) media, which began to solidify. The double layer medium was allowed to stand for about 30 minutes and was then spotted with several bacteriophage samples, which were tested to determine the host range. Each bacteriophage was spotted with as much as 2.5 µl.
Bacteriophage propagation
The propagation of bacteriophage particles was carried out using the spot test method [15]. The spot test was carried out on solidified medium by dripping 5 µl of bacteriophage particles on double-layer top agar medium. The medium that had been treated with the spot test was incubated for 24 hours at 37°C or room temperature. Next, the spot test results were shaken with sodium chloride, magnesium sulphate, and gelatine buffer (SM buffer), which was then incubated at 4°C overnight. The particle suspension obtained was centrifuged at 12,000 rpm at room temperature for 15 minutes. The supernatant obtained was then filtered using a Macherey-Nagel disposable 0.20 µm syringe filter [16].
Identification of nucleic acid type
The method used to determine the type of bacteriophage nucleic acid refers to Narulita et al. [17], which begins by extracting genetic material on the bacteriophage particles. Then 200 µl of the bacteriophage suspension in sodium chloride, magnesium sulphate, and gelatine buffer (SM buffer) was extracted and added to 20 µl Proteinase-K (Thermo Fisher Scientific), which is classified as a serine protease for cleavage of peptide bonds, and 280 µl lysis buffer. The solution mixture was shaken and incubated using a dry box unit at 65°C for 15 minutes. The 500 µl incubation supernatant was added to 500 µl phenol-chloroform-isopropanol, then centrifuged at 10,000 rpm at room temperature for 10 minutes. The supernatant was removed and added with phenol-chloroform-isopropanol and centrifuged at 10,000 rpm at room temperature for 10 min. The supernatant was then added to 50 µl each of sodium acetate (NaAc) (Merck) and isopropanol (Merck), and then incubated at –20°C for 3 hours. After incubation, it was centrifuged at 10,000 rpm for 15 min. The pellets were added with 50 µl of 70% EtOH then centrifuged at 10,000 rpm at room temperature for 5 minutes. The pellets were dried for approximately 45 minutes, then 500 µl of tris-EDTA (TE buffer) (Merck) was added to protect DNA or RNA from degradation, and it was then stored at –20°C.
Results
Isolation and purification of bacteriophages
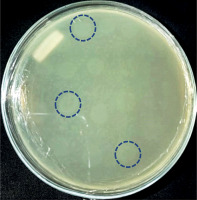
The results of bacteriophage isolation from swab samples of diabetic ulcer isolates were characterized by the formation of plaque on the surface of the double-layer top agar medium (Fig. 1).
Host range
The results of host range test of isolates AFaV1, AFaV2, AFaV3, AFaV4, AFaV5, and AFaV6 showed that they can infect bacteria other than their hosts (Table 1).
Discussion
Alcaligenes faecalis belongs to the group of gram-negative bacteria that have obligate aerobic, oxidase positive, and positive catalase properties [7]. A. faecalis is commonly found in soil, water, and hospital environments and can cause endocarditis, meningitis, skin and soft tissue infections, urinary tract infections, peritonitis, and pneumonia [7]. The presence of A. faecalis bacteria in diabetic ulcer infection was found in a recent study [6, 7]. Diabetic ulcers have been treated with antibiotics on a regular basis. However, this can lead to antibiotic resistance properties of diabetic ulcer bacteria, including Alcaligenes faecalis. Based on the latest research, the nature of XDR in A. faecalis bacteria began in 2019 with the discovery of these bacterial infections in 9 cases of skin infections in diabetic ulcers [7].
Isolation of bacteriophages with primary host bacteria of Alcaligenes showed plaque formation (Fig. 1), which indicated an infection mechanism carried out by bacteriophages against their hosts. Infection can occur because bacteriophages are able to degrade the cell wall of host bacteria through the production of the enzyme endolysin, which is encoded in the genetic material of the bacteriophage. The endolysin enzyme can hhydrolyse the peptidoglycan layer, which is followed by the occurrence of osmotic pressure disturbances in the bacterial cell wall [11, 14, 18].
Six bacteriophages isolate in this study had a diameter of ±3 mm. The diameters of the plaques formed from bacteriophages vary in size, depend on the infecting bacteriophage. The smallest plaque diameter is < 1 mm and the largest is 7 mm [19]. Plaques can be grouped into 3 sizes: small if they are less than 2 mm in size, medium-sized if they are 2 mm in size, and large if they are more than 2 mm in size [20]. A recent study on grouping bacteriophages explained that bacteriophages with plaque diameters of 3 mm can be categorized into the Siphoviridae family [21]. The Siphoviridae family is a group of bacteriophages with a capsid size of 60 nm [22]. It makes the bacteriophage of this family have the ability to diffuse faster into the host cell compared to other groups. It is also related to the size of plaque produced on the surface of the medium, which is larger than for the other groups [15, 23–25]. Based on the plaque formed, it showed that the isolates obtained in this study were lysogenic, characterized by cloudy plaques [15].
The lysogenic type bacteriophage binds to specific receptors on the cell wall of the host bacterium, injects its genetic material into the host cell, then integrates its genetic material with the genetic material of the host bacterium and is replicated together and does not control the bacterial cell so that the bacterial cell can still execute activities [26, 27]. Lysogenic bacteriophages produce cloudy plaques that can occur due to imperfect lysis of bacterial cells. The imperfections or imperfections that occur are related to the level of adsorption on weak host cells resulting in immature bacteriophage particles [3, 24, 25, 28, 29]. Lysogenic bacteriophages will form prophages that can reduce the productivity and growth of host bacteria [10]. Lysogenic bacteriophage under certain conditions or circumstances will be able to induce the occurrence of a lytic phase, so that it can completely lyse host cells [30, 31].
The lysogenic cycle in bacteriophages is promoted by a suppressor protein called CI. CI will bind to other genes and suppress the expression of these genes, so that the lytic cycle in bacteriophages is inhibited. The transition process in lysogenic bacteriophages is also promoted by 2 other proteins known as CII and CIII. Both proteins will bind to critical promoters and stimulate transcription by CI proteins. The stability of the CII protein is determined by the energy level in the cell. If a cell has enough energy, the concentration of lytic cycle AMP (cAMP) in the cell will be low, but when the cell lacks energy – for example, intracellular glucose in low concentrations – the concentration of cAMP will increase. Increasing the concentration of cAMP will encourage the stability of CII so that a lysogenic cycle can occur [31].
The process of lysogenic bacteriophage infection generally goes through 3 stages, i.e. establishment, maintenance, and induction of productive cycles:
the formation of a bacteriophage to enter the lysogenic cycle is determined by several things, including the genetic suitability between the host and bacteriophage, the physiological conditions of the host bacterium – for example a decrease in the nutrients possessed by the host cell will result in an increased risk of a lysogenic cycle, and the density of infecting bacteriophages, and the higher MOIs will increase the occurrence of the lysogenic cycle;
maintenance occurs after the prophage is generated from the integration process between the host’s genetic material and the bacteriophage. In this process, prophage can change cellular processes in host bacteria;
induction of the productive cycle of bacteriophages can occur due to external factors that trigger a DNA damage response in host bacterial cells. This process can inactivate CI gene expression, resulting in a lysogenic-lytic cycle. The induction process of this prophage can trigger the induction of other prophages [30, 32, 33].
Bacteriophages generally have the ability to infect bacteria within a narrow host range or are host specific [15, 24]. The results of the host range test in this study showed that the 6 isolates could infect bacteria other than their host, so it can be said that the 6 isolates had a wide host range. AFaV1, AFaV2, and AFaV4 isolates could infect all bacteria used, namely Klebsiella pneumoniae, Escherichia coli, and Staphylococcus aureus. Meanwhile, bacteriophage isolates AFaV3, AFaV5, and AFaV6 can infect Klebsiella pneumoniae and Staphylococcus aureus bacteria (Table 1). Bacteriophages have a broad infectious ability or have a wide host range because these bacteriophages have many protein binding receptors on their capsid, thus providing the ability of at least 2 proteins to adsorb and recognize different structures in the infected host bacteria. Bacteriophages with broad infectious capabilities have the advantage that they can kill more than one type of bacteria when different infections occur, compared to specific bacteriophages that have limited infection, which can only infect one type of bacteria [24, 34]. Bacteriophages with a broad spectrum are commonly used for phage therapy. These bacteriophages belong to the order Caudovirales, Family Myoviridae, Siphoviridae, and Podoviridae [35].
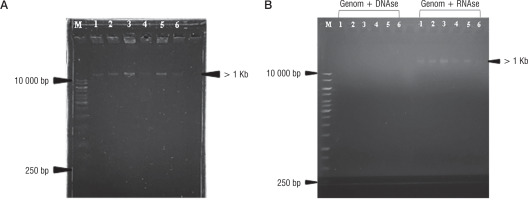
Nucleic acid extraction results of bacteriophage isolates AFaV1, AFaV2, AFaV3, AFaV4, AFaV5, and AFaV6 were visualized using a UV Transilluminator (Fig. 2). It is known that the 6 bacteriophage isolates that have been isolated have genome lengths of more than 1Kb or 10,000 base pairs. Bacteriophages generally have the nucleic acid type DNA or RNA. The nucleic acid possessed by bacteriophages can be one of the factors in the classification of bacteriophages, so it is necessary to test it in order to determine the type of nucleic acid possessed by bacteriophages that have been isolated [34]. The type of nucleic acid can be determined by treating the DNAse and RNAse enzymes to the isolated sample. DNAse enzymes are enzymes that play a role in the DNA degradation process, while RNAse enzymes play a role in RNA degradation [36]. The 6 isolated bacteriophage samples were known to have DNA nucleic acids. It can be seen from the extraction of samples that are degraded by the DNAse enzyme but not degraded by the RNAse enzyme (Fig. 2B). It is also known that bacteriophages belonging to the Siphoviridae family have DNA nucleic acids [36].
Conclusions
The type of nucleic acid possessed by 10 isolates was dsDNA, and they belonged to the Siphoviridae family group. AFaV1, AFaV2, AFaV3, AFaV4, AFaV5, and AFaV6 isolates can be said to have a wide range because they can infect bacteria other than their hosts. AFaV1, AFaV2, and AFaV4 isolates could infect all bacteria used, while AFaV3, AFaV5, and AFaV6 bacteriophage isolates could infect Klebsiella pneumoniae and Staphylococcus aureus. The 6 isolates that were isolated had the ability to infect by forming a prophage that could inhibit the growth of Alcaligenes faecalis.

 ENGLISH
ENGLISH